Characterization of “Brachyspira hampsonii” clades I and II isolated from commercial swine in Western Canada
Abstract
A novel Brachyspira emerged in 2009 and has since become a production-limiting pathogen of pigs in North America. The name “Brachyspira hampsonii” has been proposed for this novel taxon. “Brachyspira hampsonii” is divided into two phylogenetically distinct clades based on the sequence of the NADH-oxidase (nox) gene, although the clinical disease associated with clades I and II is indistinguishable and phenotypic characteristics that discriminate the clades have not been determined. The objectives of the current study were to enhance the description of the provisional species “B. hampsonii” with biochemical profiles and morphometric data from isolates affecting Canadian swine and to investigate potentially diagnostically informative characteristics for this emerging pathogen. Biochemical profiles of isolates from different commercial swine barns in Western Canada showed that biochemical profiles were insufficient to distinguish “B. hampsonii” clades I and II from each other or from other pathogenic Brachyspira. Hippurate hydrolysis, previously reported as uniformly negative in “B. hampsonii,” was variable among Canadian isolates. Spirochete dimensions and flagella numbers for “B. hampsonii” overlapped with other Brachyspira species. Taken together, these results indicate that nox gene sequencing remains a preferred method for identification and discrimination of “B. hampsonii” from other pig-associated Brachyspira spp.
Introduction
The genus Brachyspira consists of Gram-negative, motile, β-haemolytic, aerotolerant spirochaetes infecting the gastrointestinal tracts of wild and domestic animals, as well as humans. In swine, Brachyspira hyodysenteriae is the causative agent of swine dysentery (Harris et al. 1972), whereas Brachyspira pilosicoli infection is associated with spirochaetal colitis (Trott et al. 1996). “Brachyspira suanatina”, originally isolated from mallard ducks, has also been shown to cause dysentery-like disease in experimentally inoculated pigs (Råsbäck et al. 2007). Other species (Brachyspira intermedia, Brachyspira innocens, and Brachyspira murdochii) detected in pigs are non-pathogenic or have been reported to cause mild catarrhal colitis under experimental conditions (Stanton et al. 1997; Jensen et al. 2010). Re-emergence of Brachyspira-associated mucohaemorrhagic diarrhea in swine, in Western Canada and the USA, has been associated with a novel taxon (Harding et al. 2010), provisionally named “Brachyspira hampsonii” (Chander et al. 2012). “Brachyspira hampsonii” has also recently been detected in birds in Spain, in pigs in the Czech Republic, and in pigs transported to Germany from Belgium (Martinez-Lobo et al. 2013; Mahu et al. 2014; Rohde et al. 2014). Phylogenetic analysis of the NADH-oxidase (nox), 16S rRNA, and 23S rRNA genes revealed two distinct clades within the taxon (Chander et al. 2012; Burrough et al. 2013; Rubin et al. 2013b). Replacement of clades I and II with genetic groups 1–4 has also been suggested based on multi-locus sequence typing of six targets for “B. hampsonii” (Mirajkar et al. 2015). Although the clinical disease associated with clades I and II is indistinguishable (Rubin et al. 2013a; Costa et al. 2014), the phenotypic properties distinguishing them have not been established.
Brachyspira do not form visible colonies on agar, so growth is detected on blood agar medium by observing zones of β-haemolysis. Phenotypic characterization of Brachyspira is generally based on the evaluation of the strength of β-haemolysis on blood agar and activity of select biochemical markers (hippurate hydrolysis, indole production, α-galactosidase, α-glucosidase, and β-glucosidase). Spirochaete size and number of periplasmic flagella can also be used to discriminate species, although these approaches are not practical for the diagnostic laboratory. The phenotypic and genotypic characteristics of “B. hampsonii” isolates in the USA have been described (Chander et al. 2012; Mirajkar et al. 2015). Here, we describe phenotypic characteristics of “B. hampsonii” clade I and clade II isolates collected from clinical cases of mucohaemorrhagic diarrhea in Western Canadian swine between 2009 and 2013. The objectives of this study were to enhance the description of the provisional species “B. hampsonii” with information from isolates affecting Canadian swine and to investigate potentially diagnostically informative characteristics for this emerging pathogen.
Materials and methods
Selection of clinical isolates
“Brachyspira hampsonii” clade I isolate D11-30599 and clade II isolate D09-30446 were the first Canadian isolates of each clade derived from clinical cases of mucohaemorrhagic diarrhea in 2011 and 2009, respectively. The other isolates included in the study (one additional clade I and seven additional clade II) were selected from a collection of isolates from 443 “B. hampsonii”-confirmed clinical cases from different swine production sites in Alberta, Manitoba, and Saskatchewan between 2009 and 2013. The original identification of the isolates from these clinical cases was done using culture, nox sequencing, and clade-specific PCR with previously described assays (Rubin et al. 2013a; Costa et al. 2014). Although a number of clade I and clade II isolates were obtained from farms during this time period, isolates were selected for the current study only if they originated from different production systems (i.e., were epidemiologically distinct). Brachyspira isolates from clinical cases were stored long-term at −80 °C in brain-heart infusion (BHI) broth containing 10% (v/v) glycerol.
nox gene sequencing and phylogenetic analysis
Genomic DNA was extracted and purified from Joe’s Brachyspira Sask30446 (JBS) broth cultures (BHI with 1% (w/v) glucose, 5% (v/v) fetal bovine serum, 5% (v/v) sheep blood; Becton Dickinson Canada, Mississauga, Ont.) using DNeasy Blood & Tissue Kit (Qiagen Inc., Mississauga, Ont.) following the manufacturer’s instructions for pre-treatment for Gram-negative bacteria and the Animal Tissue protocol. A 939-bp region of the nox gene was amplified using genus-specific primers Bnoxf (5′-TAG CYT GCG GTA TYG CWC TTT GG-3′) and Bnoxr (5′-CTT CAG ACC AYC CAG TAG AAG CC-3′) as previously described (Rohde et al. 2002; Rubin et al. 2013a). PCR products were purified using the EZ-10 Spin Column PCR Products Purification Kit (Bio Basic Inc., Markham, Ont.) and sequenced with the amplification primers. Raw sequence data were trimmed to a uniform length of 810 bp prior to alignment with ClustalW and phylogenetic tree construction using PHYLIP (Felsenstein 1989).
Biochemical tests
Study isolates were grown in BHIS (BHI supplemented with 1% (w/v) glucose and 10% (v/v) fetal bovine serum) broth to mid-log phase. Cultures were adjusted to McFarland 5 and enzymatic activities were determined using the API ZYM kit (bioMérieux, St. Laurent, QC) according to the manufacturer’s instructions. Spot indole and hippurate broth assays were performed as previously described (Fellström and Gunnarsson 1995). Brachyspira hyodysenteriae ATCC 27164T and B. pilosicoli ATCC 51139T were included as controls in all biochemical tests.
Electron microscopy
Log phase Brachyspira cultures were prepared for transmission electron microscopy (TEM) using the single droplet negative staining technique and carbon-coated 200 mesh-200G-CP grids (Trott et al. 1996; Stanton et al. 1998). Flagella counts, cell width (average of three locations along the length of the spirochaete not including the tapered region near both ends), and cell length were measured using Image-Pro Plus (Media Cybernetics, Rockville, Md.) on captured TEM images. Twenty cells of each isolate were measured. Images were considered suitable for width and length measurements if the cells were intact, isolated, and not twisted on themselves. Flagella counts were performed on untwisted cells with at least one end visible.
Results and discussion
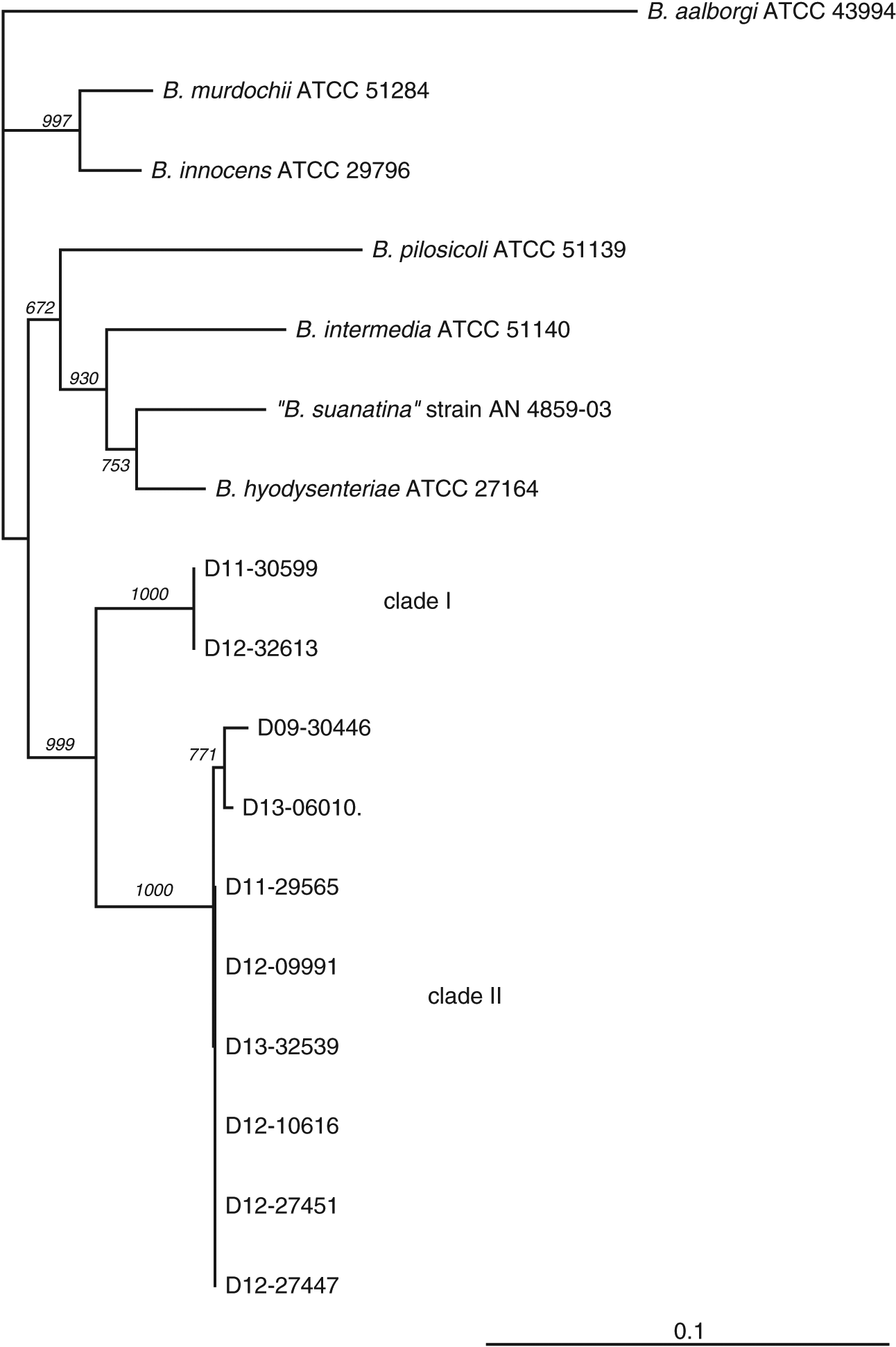
Clinical isolates of “B. hampsonii” were selected to represent distinct pig production sites, although most were related by a common source of replacement females. The number of isolates included in the study from either clade (two clade I and eight clade II) reflects higher isolation rates of clade II isolates from clinical samples submitted to our laboratory, which to our knowledge was the only laboratory performing clade level identification of “B. hampsonii” in Western Canada during the period of study. This trend in relative occurrence of clades I and II “B. hampsonii” is opposite to that reported in a recent study of “B. hampsonii” diversity in the USA (Mirajkar et al. 2015). Clade affiliation was confirmed, and relatedness of the selected clinical isolates to other Brachyspira species was investigated based on partial nox gene sequences. The nox sequences of the two clade I isolates were identical, whereas the pairwise identities between clade II isolates ranged from 99% to 100% (Fig. 1).
Fig. 1.
Results of the API ZYM, indole production, and hippurate hydrolysis tests for study isolates are summarized in Table 1. Activities of α-galactosidase, α-glucosidase, and β-glucosidase, and indole production and hippurate hydrolysis have been utilized as biochemical markers for differentiation of Brachyspira species (Achacha and Messier 1991; Fellström and Gunnarsson 1995). All clade I and clade II study isolates were negative for α-galactosidase activity, one clade II isolate was positive for α-glucosidase, and both clade I isolates and 6/8 clade II isolates were positive for β-glucosidase activity (Table 1). Although indole production was negative for all isolates as expected, hippurate hydrolysis activity was inconsistent among isolates in both clades (Table 1). These results are consistent with a previous phenotypic description of “B. hampsonii” from the USA (Chander et al. 2012) in that none of these assays could distinguish clade I and clade II isolates. Interestingly, all isolates in the USA study (n = 17 clade I, n = 3 clade II) were negative for hippurate hydrolysis activity (Chander et al. 2012). Variations in indole production, hippurate hydrolysis, α-galactosidase, and α-glucosidase activities of human isolates of B. pilosicoli have been observed in previous studies (De Smet et al. 1998), and variation in indole production of swine-isolated B. hyodysenteriae has also been reported (Fellström et al. 1999). The greater consistency in hippurate hydrolysis activity in the USA study may reflect epidemiological relationships of isolates, but these relationships are not described in that report. Taken together, these results show that none of these biochemical assays can discriminate clades I and II or “B. hampsonii” from B. hyodysenteriae (Table 2). The lack of specificity of biochemical markers for differentiating Brachyspira species has inspired the development of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) as a promising phenotypic method for differentiation of Brachyspira. This technology has been demonstrated to distinguish Brachyspira species (Calderaro et al. 2013; Prohaska et al. 2014), but clades of “B. hampsonii” are not reliably distinguished (Warneke et al. 2014). The initial capital costs for diagnostic MALDI-TOF have also limited its widespread adoption.
Table 1.
| Clade Ia | Clade IIa | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| D11-30599 | D12-32613 | D09-30446 | D11-29565-A6P8 | D12-09991-28 | D12-10616-P8 | D12-27447 | D12-27451 | D12-32539 | D13-06010 | |
| Alkaline phosphatase | + | W+b | + | + | + | + | − | W+ | + | − |
| Esterase (C4) | + | + | + | + | − | + | + | + | + | − |
| Esterase lipase (C8) | + | W+ | + | + | − | + | + | W+ | + | − |
| Lipase (C14) | − | − | − | − | − | − | − | − | − | − |
| Leucine arylamidase | − | − | − | − | − | − | − | − | − | − |
| Valine arylamidase | − | − | − | − | − | − | − | − | − | − |
| Cystine arylamidase | − | − | − | − | − | − | − | − | − | − |
| Trypsin | − | − | − | − | − | − | − | − | − | − |
| α-Chymotrypsin | − | − | W+ | − | − | − | − | − | − | − |
| Acid phosphatase | + | + | + | + | W+ | + | + | + | + | W+ |
| Naphthol-AS-BI-phosphohydrolase | + | + | W+ | + | W+ | + | + | + | + | + |
| α-Galactosidase | − | − | − | − | − | − | − | − | − | − |
| β-Galactosidase | + | + | + | + | W+ | + | + | + | + | − |
| β-Glucuronidase | − | − | − | − | − | − | − | + | − | − |
| α-Glucosidase | − | − | − | − | − | − | − | − | − | + |
| β-Glucosidase | + | + | + | + | − | + | W+ | + | + | − |
| N-acetyl-β-glucosaminidase | − | − | − | − | − | − | − | − | − | − |
| α-Mannosidase | − | − | − | − | − | − | − | − | − | − |
| α-Fucosidase | − | − | − | − | − | − | − | − | − | − |
| Indole production | − | − | − | − | − | − | − | − | − | − |
| Hippurate hydrolysis | − | + | − | + | − | + | + | + | + | − |
a
Based on the partial nox gene sequence.
b
W+ denotes a weak positive.
Table 2.
| Characteristic | “B. Hampsonii” clade I | “B. Hampsonii” clade II | B. hyodysenteriae (Trott et al. 1996; Fellström et al. 1999) | “B. suanatina” (Råsbäck et al. 2007; Mushtaq et al. 2015) | B. pilosicoli (Trott et al. 1996) | B. innocens (Oxberry et al. 1998) | B. murdochii (Stanton et al. 1997; Jensen et al. 2010) | B. intermedia (Stanton et al. 1997; Jensen et al. 2010) |
|---|---|---|---|---|---|---|---|---|
| Cell length (μm) | 7.27 ± 1.30a | 8.53 ± 2.19a | 9.78 ± 1.87c | 7.27 ± 1.00c | 6.27 ± 0.98c | 9.40 ± 1.85c | 5–8d | 7.5–10d |
| Cell diameter (μm) | 0.26 ± 0.045a | 0.46 ± 0.05a | 0.35 ± 0.02c | 0.3 ± 0.00e | 0.27 ± 0.03c | 0.36 ± 0.03c | 0.35–0.40d | 0.35–0.45d |
| No. of flagella | 11–16b | 12–15b | 8–12 | 14–16 | 4–6 | 10–13 | 11–13 | 12–14 |
| β-Haemolysis | Strong | Strong | Strong | Strong | Weak | Weak | Weak | Weak |
| Indole production | − | − | Variable | Weak+ / +f | − | − | − | + |
| Hippurate hydrolysis | Variable | Variable | − | − | + | − | − | − |
| α-Galactosidase | − | − | − | − | + | + | − | − |
| α-Glucosidase | − | Variable | + | Weak+ | − | − | − | + |
| β-Glucosidase | + | Variable | + | + | − | + | − | + |
a
Mean ± standard deviation for 20 spirochaetes.
b
Range for 15 spirochaetes.
c
Mean ± standard deviation for 5 spirochaetes.
d
Authors’ reported range.
e
Mean ± standard deviation for 10+ spirochaetes.
f
Reported as positive by Mushtaq et al. (2015) and weak positive by Råsbäck et al. (2007).
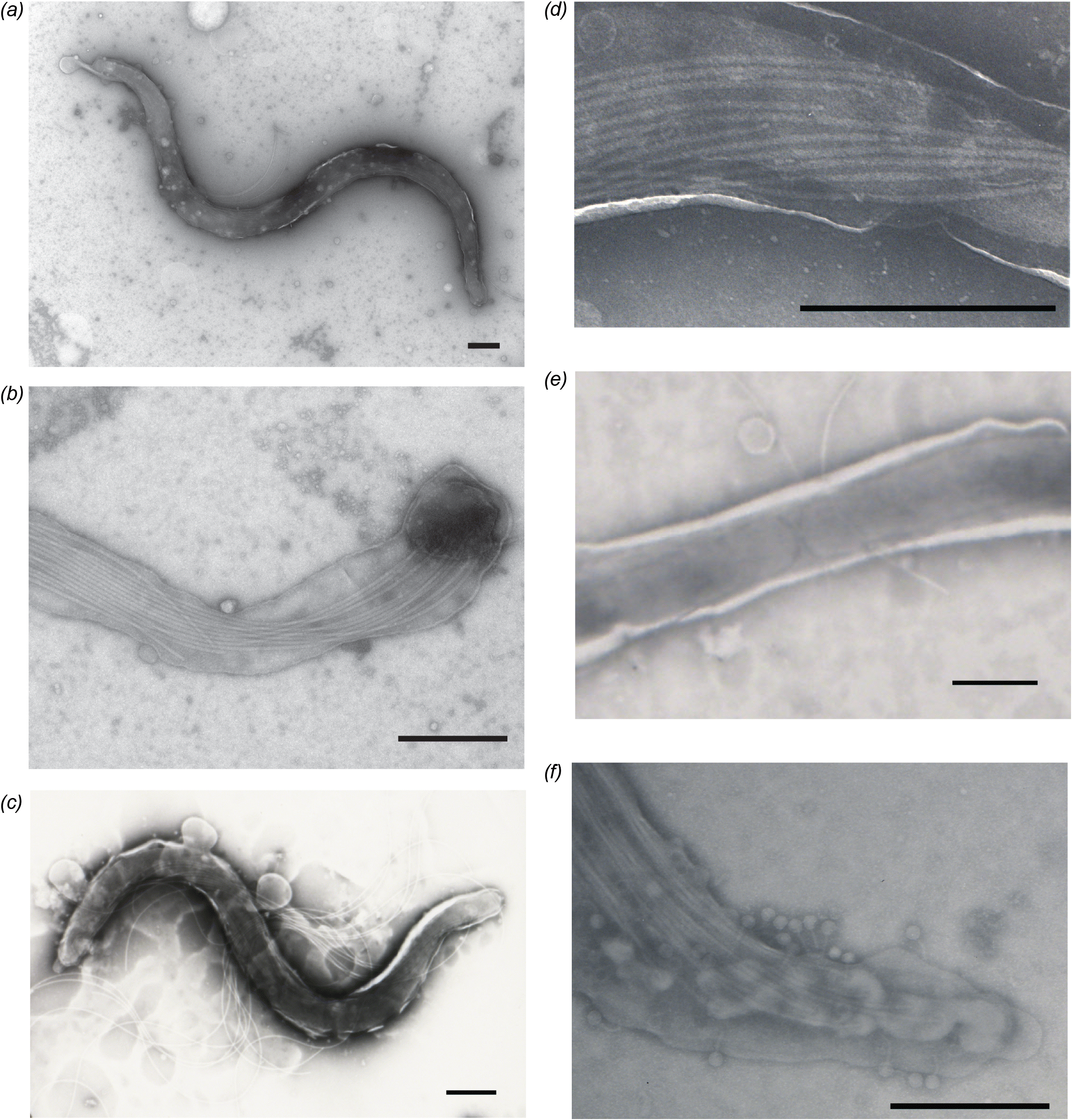
TEM was performed on D11-30599 and D09-30446 to measure the dimensions of the spirochaetes and count the number of flagella for comparison to other Brachyspira species. Representative images are shown in Fig. 2. Spirochaetes of clade I “B. hampsonii” isolate D11-30599 were 7.27 ± 1.30 μm and 0.26 ± 0.045 μm, with 11–16 periplasmic flagella (Figs. 2a, 2b and Table 2). For clade II isolate D09-30446, average length and width were 8.53 ± 2.19 μm and 0.46 ± 0.05 μm, with 12–15 periplasmic flagella (Figs. 2c, 2d and Table 2). Apparently dividing cells were observed (Fig. 2e), and tailed bacteriophages were occasionally detected on the surface D09-30446 spirochaetes under TEM (Fig. 2f). The presence of tailed bacteriophages is consistent with the type of bacteriophage detected with other Brachyspira (Calderaro et al. 1998; Ackermann 2007). The morphological measurements and flagella counts for “B. hampsonii” overlapped with other pig-associated Brachyspira species and so do not on their own provide species identification (Table 2).
Fig. 2.
Conclusions
The biochemical variation of “B. hampsonii” isolates from Western Canadian swine production systems highlights that biochemical characterization alone is not diagnostically informative. Although some traits of “B. hampsonii” isolates are consistent (indole and α-galactosidase negative), these characteristics are not solely sufficient to discern them from other Brachyspira species as previously observed in the USA study (Chander et al. 2012). The morphology of selected isolates was also indistinguishable from other species of Brachyspira. Based on these results and previous observations, nox gene sequencing remains a useful, accessible, and affordable tool for identification and discrimination of Brachyspira spp. affecting pigs.
Acknowledgements
The authors thank Ian Shirley, Chris Stuart, and Samantha Ekanayake (Western College of Veterinary Medicine) for their expertise in electron microscopy. This project was supported by funding from the Canadian Swine Health Board.
References
Achacha M, and Messier S. 1991. Identification of Treponema hyodysenteriae and Treponema innocens using two four-hour identification systems. Journal of Veterinary Diagnostic Investigation, 3: 211–214.
Ackermann HW. 2007. 5500 Phages examined in the electron microscope. Archives of Virology, 152: 227–243.
Burrough ER, Wilberts BL, Bower LP, Jergens AE, and Schwartz KJ. 2013. Fluorescent in situ hybridization for detection of “Brachyspira hampsonii” in porcine colonic tissues. Journal of Veterinary Diagnostic Investigation, 25: 407–412.
Calderaro A, Dettori G, Collini L, Ragni P, Grillo R, Cattani P, et al. 1998. Bacteriophages induced from weakly beta-haemolytic human intestinal spirochaetes by mitomycin C. Journal of Basic Microbiology, 38: 323–335.
Calderaro A, Piccolo G, Montecchini S, Buttrini M, Gorrini C, Rossi S, et al. 2013. MALDI-TOF MS analysis of human and animal Brachyspira species and benefits of database extension. Journal of Proteomics, 78: 273–280.
Chander Y, Primus A, Oliveira S, and Gebhart CJ. 2012. Phenotypic and molecular characterization of a novel strongly hemolytic Brachyspira species, provisionally designated “Brachyspira hampsonii”. Journal of Veterinary Diagnostic Investigation, 24: 903–910.
Costa MO, Hill JE, Fernando C, Lemieux HD, Detmer SE, Rubin JE, et al. 2014. Confirmation that “Brachyspira hampsonii” clade I (Canadian strain 30599) causes mucohemorrhagic diarrhea and colitis in experimentally infected pigs. BMC Veterinary Research, 10: 129.
De Smet KA, Worth DE, and Barrett SP. 1998. Variation amongst human isolates of Brachyspira (Serpulina) pilosicoli based on biochemical characterization and 16S rRNA gene sequencing. International Journal of Systematic Bacteriology, 48(4): 1257–1263.
Fellström C, and Gunnarsson A. 1995. Phenotypical characterisation of intestinal spirochaetes isolated from pigs. Research in Veterinary Science, 59: 1–4.
Fellström C, Karlsson M, Pettersson B, Zimmerman U, Gunnarsson A, and Aspan A. 1999. Emended descriptions of indole negative and indole positive isolates of Brachyspira (Serpulina) hyodysenteriae. Veterinary Microbiology, 70: 225–238.
Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics, 5: 164–166.
Harding J, Hill JE, Chirino-Trejo M, O’Connor B, Rubin J, and Fernando C. 2010. Detection of a novel Brachyspira species associated with haemorrhagic and necrotizing colitis. In Proceedings of the 21st Annual International Pig Veterinary Society Congress, Vancouver, BC, 18–21 July 2010. pp. 1–5.
Harris DL, Glock RD, Christensen CR, and Kinyon JM. 1972. Inoculation of pigs with Treponema hyodysenteriae (new species) and reproduction of the disease. Veterinary Medicine Small Animal Clinician, 67: 61–64.
Jensen TK, Christensen AS, and Boye M. 2010. Brachyspira murdochii colitis in pigs. Veterinary Pathology, 47: 334–338.
Mahu M, De Jong E, De Pauw N, Vande Maele L, Vandenbroucke V, Vandersmissen T, et al. 2014. First isolation of “Brachyspira hampsonii” from pigs in Europe. Veterinary Record, 174: 47.
Martinez-Lobo FJ, Hidalgo A, Garcia M, Argüello H, Naharro G, Carvajal A, et al. 2013. First identification of “Brachyspira hampsonii” in wild European waterfowl. PLoS ONE, 8: E82626.
Mirajkar NS, Bekele AZ, Chander YY, and Gebhart CJ. 2015. Molecular epidemiology of novel pathogen “Brachyspira hampsonii” reveals relationships between diverse genetic groups, regions, host species, and other pathogenic and commensal Brachyspira species. Journal of Clinical Microbiology, 53: 2908–2918.
Mushtaq M, Zubair S, Råsbäck T, Bongcam-Rudloff E, and Jansson DS. 2015. Brachyspira suanatina sp. nov., an enteropathogenic intestinal spirochaete isolated from pigs and mallards: genomic and phenotypic characteristics. BMC Microbiology, 15: 208.
Oxberry SL, Trott DJ, and Hampson DJ. 1998. Serpulina pilosicoli, waterbirds and water: potential sources of infection for humans and other animals. Epidemiology and Infection, 121: 219–225.
Prohaska S, Plfüger V, Ziegler D, Scherrer S, Frei D, Lehmann A, et al. 2014. MALDI-TOF MS for identification of porcine Brachyspira species. Letters in Applied Microbiology, 58: 292–298.
Råsbäck T, Jansson DS, Johansson KE, and Fellström C. 2007. A novel enteropathogenic, strongly haemolytic spirochaete isolated from pig and mallard, provisionally designated ‘Brachyspira suanatina’ sp. nov. Environmental Microbiology, 9: 983–991.
Rohde J, Habighorst-Blome K, and Seehusen F. 2014. “Brachyspira hampsonii” clade I isolated from Belgian pigs imported to Germany. Veterinary Microbiology, 168: 432–435.
Rohde J, Rothkamp A, and Gerlach GF. 2002. Differentiation of porcine Brachyspira species by a novel nox PCR-based restriction fragment length polymorphism analysis. Journal of Clinical Microbiology, 40: 2598–2600.
Rubin JE, Costa MO, Hill JE, Kittrell HE, Fernando C, Huang Y, et al. 2013a. Reproduction of mucohaemorrhagic diarrhea and colitis indistinguishable from swine dysentery following experimental inoculation with “Brachyspira hampsonii” strain 30446. PLoS ONE, 8: E57146.
Rubin JE, Harms NJ, Fernando C, Soos C, Detmer SE, Harding JCS, et al. 2013b. Isolation and characterization of Brachyspira spp. including “Brachyspira hampsonii” from lesser snow geese (Chen caerulescens caerulescens) in the Canadian Arctic. Microbial Ecology, 66: 813–822.
Stanton TB, Fournié-Amazouz E, Postic D, Trott DJ, Grimont PA, Baranton G, et al. 1997. Recognition of two new species of intestinal spirochetes: Serpulina intermedia sp. nov. and Serpulina murdochii sp. nov. International Journal of Systematic Bacteriology, 47: 1007–1012.
Stanton TB, Postic D, and Jensen NS. 1998. Serpulina alvinipulli sp. nov., a new Serpulina species that is enteropathogenic for chickens. International Journal of Systematic Bacteriology, 48(3): 669–676.
Trott DJ, Atyeo RF, Lee JI, Swayne DA, Stoutenburg JW, and Hampson DJ. 1996. Genetic relatedness amongst intestinal spirochaetes isolated from rats and birds. Letters in Applied Microbiology, 23: 431–436.
Warneke HL, Kinyon JM, Bower LP, Burrough ER, and Frana TS. 2014. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for rapid identification of Brachyspira species isolated from swine, including the newly described “Brachyspira hampsonii”. Journal of Veterinary Diagnostic Investigation, 26: 635–639.
Information & Authors
Information
Published In

FACETS
Volume 1 • Number 1 • January 2017
Pages: 163 - 172
Editor: Alexandre Poulain
History
Received: 27 April 2016
Accepted: 21 July 2016
Version of record online: 19 October 2016
Copyright
© 2016 Perez et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
JER, JCSH, and JEH conceived and designed the study.
JBDSP, JER, and CF performed the experiments/collected the data.
All analyzed and interpreted the data.
All drafted or revised the manuscript.
Competing Interests
The University of Saskatchewan has filed patent applications pertaining to the “B. hampsonii” clade II strain (Brachyspira sp. SASK30446) titled “Isolated Brachyspira and methods and compositions for expanding and isolating Brachyspira” in Canada (serial number 2842068) and USA (serial number 14/233414) on 18 July 2012. The applications are published and available online at: http://patentscope.wipo.int.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Jason Byron D.S. Perez, Joseph E. Rubin, Champika Fernando, John C.S. Harding, and Janet E. Hill. 2017. Characterization of “Brachyspira hampsonii” clades I and II isolated from commercial swine in Western Canada. FACETS.
1(): 163-172. https://doi.org/10.1139/facets-2016-0018
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Characterization of Brachyspira communities from clinical cases of swine mucohaemorrhagic diarrhea through deep sequencing of the NADH oxidase ( nox ) gene
2. In vitro attenuation of a virulent swine isolate of Brachyspira hampsonii
