Introduction
Ectoparasitic arthropods have challenged terrestrial agricultural production for decades and pose serious human health risks as vectors of disease. In aquatic habitats they are significant pests to wild and farmed fishes worldwide. Parasitic copepods, and in particular those from the Family Caligidae (known as sea lice) such as
Lepeophtheirus salmonis, cost the aquaculture industry hundreds of millions of dollars annually due to treatment costs and reduced growth or value of fish (
Costello 2009). There are additional consequences of infection treatments, such as increased chemical inputs into the environment (
Burridge et al. 2010;
Veldhoen et al. 2012). Recent reviews highlight the impact of lice on salmon (
Fast 2014), the emergence of drug resistance in sea lice (
Aaen et al. 2015;
McNair 2015), and sea lice biology and control strategies in aquaculture (
Torrissen et al. 2013).
Variation in host response to sea lice infection is common inter- and intra-specifically. Among species, strong inflammatory and cellular immune responses are associated with reduced infection intensities in coho salmon (
Oncorhynchus kisutch) and pink salmon (
Oncorhynchus gorbuscha) (
Johnson and Albright 1992;
Jones et al. 2007;
Braden et al. 2012,
2015;
Sutherland et al. 2014). Within species, heritable differences in susceptibility indicate potential for selective breeding (
Kolstad et al. 2005;
Gjerde et al. 2011). Outcomes of infection differ depending on these and other factors, such as host life stage (
Jones et al. 2008b;
Sutherland et al. 2011). Infection outcomes can include growth reduction, behavioral changes, osmotic imbalance, stress response, and others, even mortality (
Pike and Wadsworth 1999;
Wagner et al. 2008;
Fast 2014). Improved understanding of the mechanisms and pathways underlying host resistance is important for effective breeding (
Yáñez et al. 2014), for predicting effects of infection in wild populations, and for tailoring immunostimulant diets against specific infections (
Alvarez-Pellitero 2008). The use of targeted functional feeds has become increasingly important in aquaculture, including in-feed immunostimulants (
Jensen et al. 2015). Approaches using in-feed immunostimulation are favoured as these can be administered without handling stress and are easily integrated into management practices, are active against multiple life stages, and are not likely to result in resistance development (
Burka et al. 1997). This process typically involves presenting the fish with a pathogen-associated molecular pattern (PAMP) and stimulating an innate immune response; for example, using peptidoglycan to stimulate antimicrobial peptide gene expression in rainbow trout (
Oncorhynchus mykiss) (
Casadei et al. 2013).
Alternative treatment options are especially needed due to recent development of multiple chemical drug resistance in sea lice (
Aaen et al. 2015;
McNair 2015). Some of these alternate approaches include biological control (cleaner fish), mechanical delousing (including sieves and tank filters), regulation improvements, and selective breeding (for review see
Torrissen et al. 2013;
Gharbi et al. 2015). Immunostimulation is another option for treatment, temporarily increasing the natural immunity of the host or directing it towards a more appropriate response type (
Bricknell and Dalmo 2005). Pulse doses of immunostimulation are used to avoid tolerance to the immunostimulant and can be used in periods of increased disease risk (
Bricknell and Dalmo 2005). Complementary approaches may be effective when applied in a treatment rotation strategy and may reduce the burden on chemical parasiticides.
Several immunostimulation approaches for salmon lice have been developed, which have used the current knowledge of appropriate salmon defenses against lice (e.g., innate mucosal responses).
Covello et al. (2012) tested the effectiveness of incorporating CpG oligodeoxynucleotide (ODN) or yeast extracts into post-smolt Atlantic salmon (
Salmo salar) feed. Salmon on the treated diet had reduced
L. salmonis infection levels (∼40% lower than controls) and showed enhanced site-specific inflammation. The difference in infection levels between control and treated fish increased over time, suggesting that the response to the feed was maintained (
Covello et al. 2012).
Purcell et al. (2013) identified that a previous exposure to lice resulted in lower reinfection density than naïvely infected salmon, and that this effect was increased further through CpG-ODN immunostimulation. Salmon with the lowest levels of lice (i.e., previously exposed and immunostimulated) had the highest expression of
il-1β (
Purcell et al. 2013). Enhanced defenses by CpG-ODN are dose dependent; at lower doses, the feed still increased
interleukin-1β (
il-1β) expression in the head kidney but did not provide as much protection as evidenced by infection levels being ∼18% lower than controls (
Poley et al. 2013). Dose-dependent host immunostimulation can augment innate immunity and local inflammation to reduce lice burdens. Other approaches have been taken to immunostimulate against salmon lice infections, although mechanisms of rejection remain unknown (see
Jensen et al. 2015).
Here, we evaluate the effects of a commercial immunostimulant diet formulation composed of bacterial cell wall extract peptidoglycan on lice rejection and on parasite transcriptome profiles using a
L. salmonis oligonucleotide microarray (
Sutherland et al. 2012;
Yasuike et al. 2012). We also evaluate the expression of several immune-related genes in the immunostimulated host using reverse transcription quantitative PCR (RT-qPCR). Characterizing the dynamics of the infection alongside molecular responses of both the parasite and the host informs on the efficacy and potential of dose-dependent immunostimulation of the hosts, as well as the effect of the stimulated host on
L. salmonis molecular physiology.
Materials and methods
Animals
Atlantic salmon smolts were obtained from Buckman Creek Hatchery, Pennfield, New Brunswick, Canada. As per Canadian federal fish health regulations for transfer of fish, a disease-free certification from a fish health professional determined this source of fish and eggs pathogen-free (i.e., free of salmonid diseases of importance). These fish were transported to the Aquatic Animal Facility in the Atlantic Veterinary College at the University of Prince Edward Island (UPEI) (Charlottetown, Prince Edward Island, Canada). All experimental protocols for the use of fish followed the guidelines provided by the
Canadian Council on Animal Care (2005;
ccac.ca/Documents/Standards/Guidelines/Fish.pdf) and were submitted for review and approval to the UPEI Animal Care Committee (UPEI Animal Care Protocol #10-014).
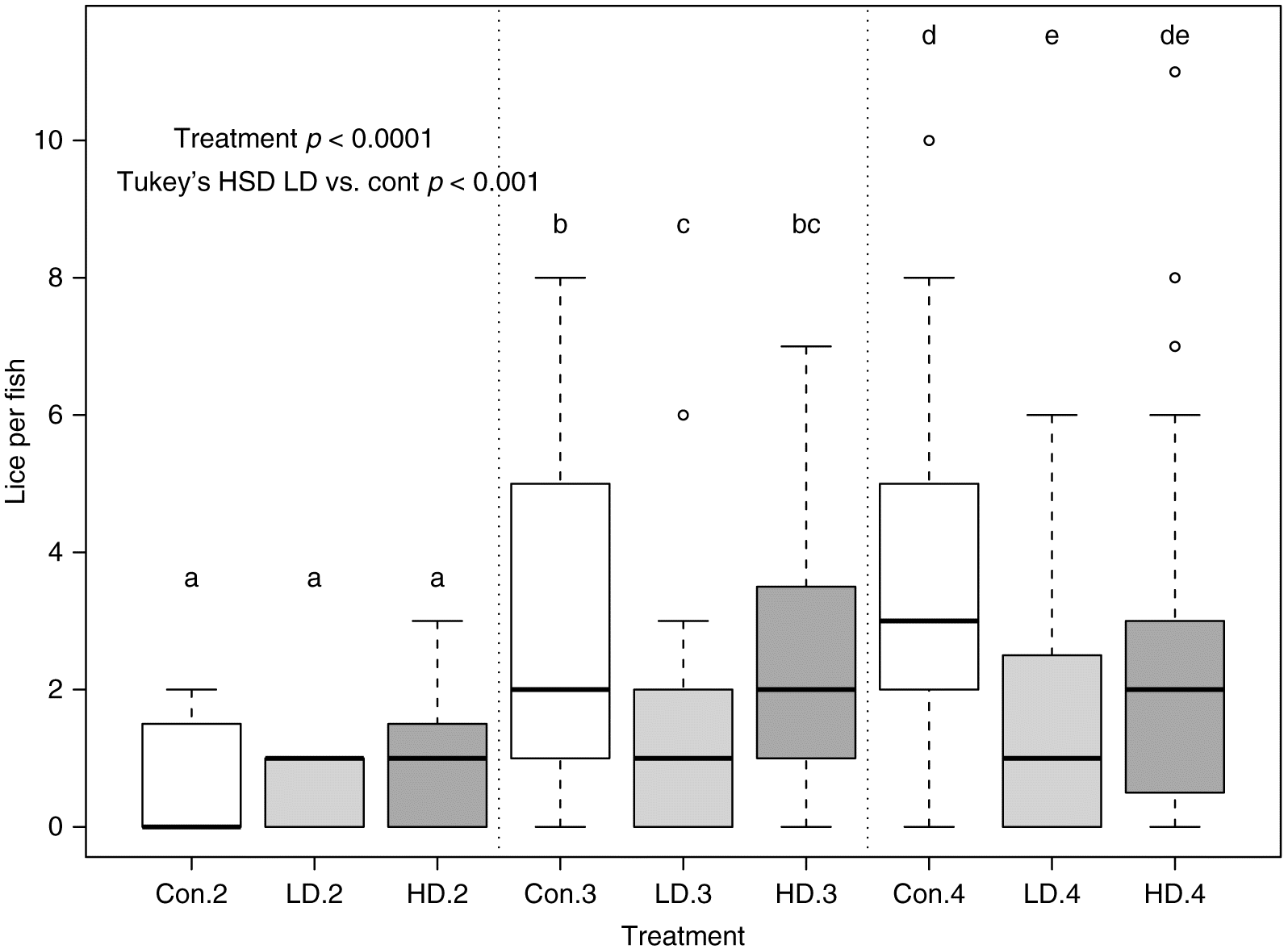
Fish (n = 40–45/tank) were held in 10 circular flow-through tanks containing 250 L of 11 °C freshwater for 3 wk to acclimate. The system was then switched to saltwater (Instant Ocean®, Cincinnati, Ohio, USA) recirculation over 7 d, resulting in a final salinity of 33 ppt. Fish were held in saltwater for a further 2 wk to acclimatize to the saltwater (11.5 °C, DO > 7.0 mg/L, pH 7.9, ammonia/nitrite/nitrate monitored). During this period, fish were fed a control diet at 1.5% body weight/day, divided over two feeds. Throughout the study, fish were kept on a light:dark cycle of 14 h:10 h. At the start of the study, fish weighed 98.2 ± 5.2 g (mean ± SEM), and at the end of the study, fish weighed 177.0 ± 10.1 g (mean ± SEM).
Egg strings from
L. salmonis were harvested from sea cage cultured Atlantic salmon (New Brunswick, Canada) from the BMA2A region (this region contains lice with elevated emamectin benzoate (EMB) resistance;
Igboeli et al. 2014) and were aerated in 13 °C saltwater collected from the sea cage site and maintained at the Atlantic Animal Facility, Atlantic Veterinary College at 33–36 salinity through 10% daily water changes until nauplii hatched and molted to copepodids (∼6–8 d).
Feed production
All feeds were produced at the EWOS Innovation facilities in Dirdal, Norway. The three different feed treatments were control/base feed (14% fat, 43% protein, maximum 3% fiber), low dose (LD; 0.12% peptidoglycan extract from bacterial cell wall with nucleotide formulation milled in with the base feed), and high dose (HD; 0.20% peptidoglycan extract from bacterial cell wall with nucleotide formulation milled in with the base feed).
Lice exposures and sampling
Fish were infected following the method of
Covello et al. (2012). Following seawater acclimatization, treatment tanks (total = 9 tanks) were randomly assigned to one of three treatment groups and placed on one of the following three diets: control/base feed, LD, or HD. A system control tank was maintained on the recirculation loop but was separated from the other tanks by a plastic barrier. In this tank, the fish were held at the same density as the other tanks (40–45/tank) and fed the control/base feed but were not exposed to lice.
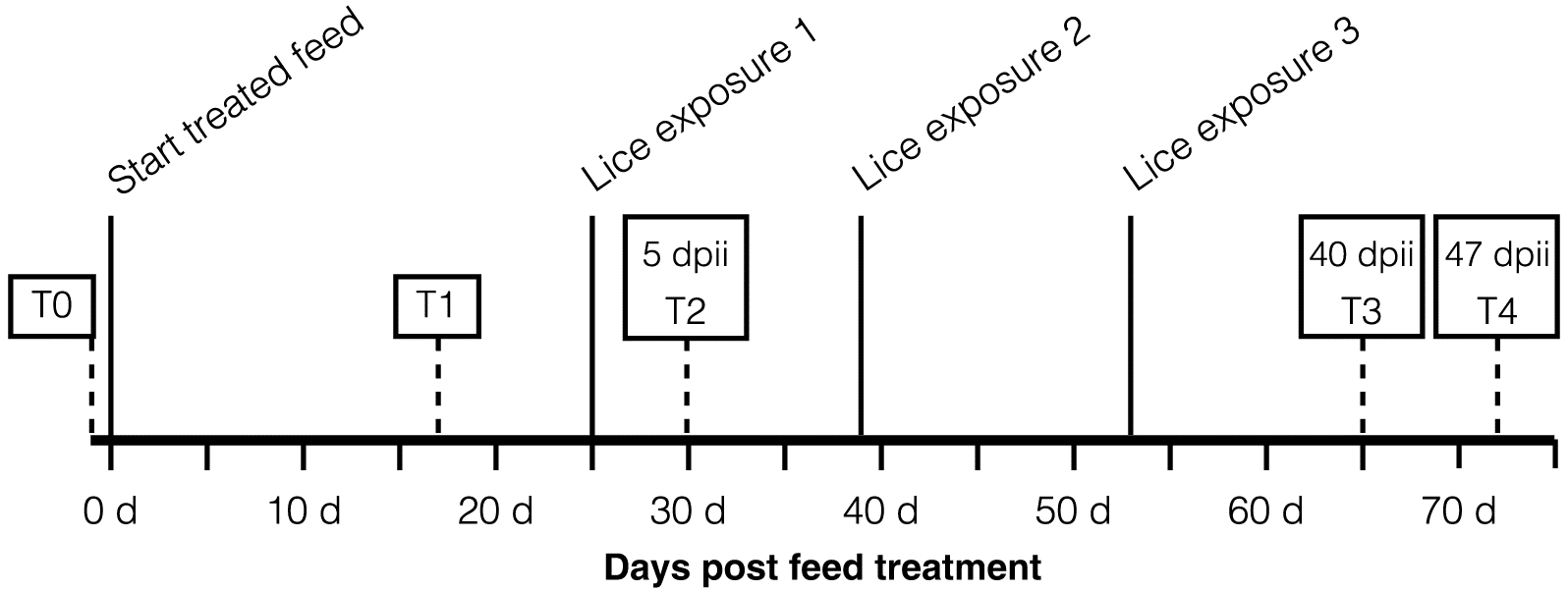
The infection and sampling regime is shown in
Fig. 1. The day prior to feed adjustment, two fish from each tank were sampled and used as a time control (T0) for reference (
n = 20 fish). For all following sampling times, six fish were sampled per tank (
n = 18 fish per diet group). Fish were maintained on treated feed for 17 d and then sampled (T1). Eight days after T1, fish in all experimental tanks were exposed to
L. salmonis copepodids at 10–12 lice/fish (initial exposure). Five days after the initial exposure, fish were sampled (T2). A second lice exposure and a third lice exposure (also at 10–12 lice/fish) were conducted 14 and 28 d, respectively, after the first exposure to mimic a more constant low-level exposure as expected in the field. Fish were sampled again at 40 and 47 d post initial infection (dpii; T3 and T4, respectively;
Fig. 1).
At each sampling time, feed was withheld for 24 h prior to sampling, and fish were euthanized with an overdose of tricaine methanesulfonate (MS-222: 250 mg/L; Syndel Laboratories Ltd., Nanaimo, British Columbia, Canada). Lice were rapidly enumerated and staged, then flash frozen in dry ice. Spleen, skin, and head kidney were excised from each salmon individual and frozen on dry ice. In an effort to standardize the sampling, skin samples were always taken posterior to the left pectoral fin above the lateral line (as in
Covello et al. 2012). All tissues were stored at −80 °C until required for gene expression analysis.
RNA extraction
Total RNA was extracted from salmon spleen, skin, and head kidney samples, and L. salmonis whole individuals using Tri Reagent (Life Technologies, Carlsbad, California, USA) according to the manufacturer’s instructions. Approximately 50 mg of still frozen tissue was added to 1.5 mL of Tri Reagent and mechanically macerated with a homogenizer (VWR, Mississauga, Ontario, Canada). Then RNA was extracted as per the manufacturer’s instructions (Invitrogen, Carlsbad, California, USA). Resultant RNA was dissolved in 100–200 μL of molecular biology grade water and stored at −80 °C. Total RNA concentration was measured using the NanoDrop-2000 spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA). A subsample of all salmon tissue RNA extractions (10%) was tested for RNA quality using automated gel electrophoresis (Experion; Bio-Rad, Mississauga, Ontario, Canada), as per the manufacturer’s instructions. The quality of salmon tissue RNA was good (RQI > 7.0) in over 90% of the samples tested. Five micrograms of the extracted RNA was then DNase treated using a TURBO DNase-free™ kit (Ambion, Foster City, California, USA) following the manufacturer’s protocol. Lice samples were all tested using the Experion, and all used for the analysis were high quality (RQI > 7.0).
Salmon RT-qPCR
Synthesis of cDNA was performed on 1 μg of DNase-treated total RNA using the Reverse Transcription System (Promega, Madison, Wisconsin, USA) and random hexamers, according to the manufacturer’s instructions. Reverse transcriptase-free (–RT) controls were included to ensure the absence of genomic DNA. cDNA samples were stored at −20 °C until use for qPCR.
Primer sets for the three reference genes (
ef-1ab,
rps20, and
18S) and four genes of interest (
il-1β,
il-8,
toll-like receptor 9, and
matrix metalloproteinase 9) were obtained from previous literature (
Frost and Nilsen 2003;
Sutherland et al. 2012;
Supplementary Material 1). Each reaction was run in a 96-well plate (Eppendorf, Mississauga, Ontario, Canada) in 10 μL reactions containing 0.5 μmol/L of each primer, 2× GoTaq
® qPCR Master Mix (Promega), and 1 μL of cDNA (diluted 1:1 in nuclease-free water). A no template control (NTC) was included for each primer pair and remained unamplified. Amplifications were carried out in an Eppendorf Mastercycler ep realplex
2 under the following conditions: initial denaturation at 95 °C for 10 min, followed by 40 amplification cycles of 95 °C for 15 s, annealing for 15 s at 55 °C, and extension for 15 s at 72 °C. Following this amplification, melt curve analysis was performed to ensure amplification of a single product.
The gene expression results were analyzed using qBasePLUS relative quantification framework software version 2.0 (
Hellemans et al. 2007). Primer efficiencies were determined by analysis of 10-fold serial dilutions for the reference genes and 5-fold serial dilutions for the genes of interest using pooled cDNA of experimental samples and samples from a separate experiment of cohort individuals 6 h after LPS injection. Efficiencies averaged 96% for all genes across salmon tissues, and all experimental samples were within the range of the standard curve. The stability of
ef-1ab,
rps20, and
18s as reference genes was evaluated using geNORM (qBASE+ Biogazelle;
Vandesompele et al. 2002), and the geometric mean of these reference genes was used to normalize target gene transcripts. Data were normalized per group by subtracting the pretreatment means from each treatment (i.e., T(0) expression level). Log
2-transformed data were used to test for significant differences between treatment groups over exposure time using linear models and pairwise Tukey’s honest significant difference (HSD) tests in R (
R Core Team 2017).
Salmon lice microarray analysis
Total RNA was extracted from lice infecting control diet fish and LD diet fish from times T3 and T4 (
Fig. 1). These individuals were used to hybridize to an
L. salmonis 38K oligonucleotide microarray (
Sutherland et al. 2012). Samples consisted of pools of both male and female adult lice collected from the same fish (each sex in equal proportion within each sample; two to four individuals per pool). Seven pools were used from the control fish and four pools from the LD. As no clustering was viewed in initial data exploration by time point (T3 or T4) and due to low sample numbers, these two time points were merged for differential expression analysis.
Samples were processed in a random order, using 200 ng of total RNA as an input for reverse transcription and cDNA amplification to Cy5-cRNA using the Low Input Quick Amp Labeling kit as per the manufacturers’ instructions (v6.5; Agilent Technologies, Santa Clara, California, USA). A reference pool was also synthesized as Cy3-cRNA using all possible treatment conditions (time points and treatments) and equal numbers of males and females in the reference. Aliquots of the reference pool were then stored at −80 °C until hybridization. Sample (Cy5) and reference pool (Cy3) combinations (825 ng each) were hybridized to randomly ordered 4 × 38K microarrays as per manufacturer’s instructions, with the optional Stabilization and Drying Solution ozone protection step (Agilent). After washing, slides were kept dark and at low ozone (ozone ≤8 ppb), and scanned on a ScanArray Express (Perkin Elmer, Waltham, Massachusetts, USA) at 5 μm resolution. Photomultiplier tube settings were kept constant for the scanning and were optimized to obtain ∼1% saturated spots (Cy5:65; Cy3:60).
Microarray probe intensity values were quantified and flagged for quality in Imagene (v8.0; BioDiscovery, El Segundo, California, USA). These values were analyzed using
limma (
Smyth 2004;
Ritchie et al. 2007) in R (
R Core Team 2017). Probes were retained for analysis when at least three samples in any condition were ≥500 for raw fluorescence in both channels. Probes saturated in all samples were removed from analysis. All spots were background corrected using the
minimum function in
limma (
Ritchie et al. 2007). The arrays were normalized by within array
loess normalization and between array quantile normalization using the Cy3 reference channel (
Smyth and Speed 2003). Probes were tested for differential expression using linear models contrasting the LD and control treatments using eBayes fit (
Smyth 2004). Differential expression required
p ≤ 0.05 and fold change (FC) ≥ 1.5.
Salmon lice RT-qPCR validation
The same RNA used for the microarrays was used for RT-qPCR. Total RNA (1.9 μg) was reverse transcribed to cDNA using Superscript III (Invitrogen) as per the manufacturer’s instructions for oligo dT priming. A representative sample from each condition was pooled, then diluted 7-fold to be the first point on a 6-point, 5-fold dilution series used as a standard curve for primer testing. Quantitative PCR was performed on an MX3000P (Agilent) using SsoFast EvaGreen Supermix with low rox (Bio-Rad) as per manufacturer’ instructions, and with the following thermal regime: 95 °C for 30 s (1 cycle); 95 °C for 5 s, 55 °C for 20 s (40 cycles); and 95 °C for 5 s, 55 °C for 10 s then increase by half-degree increments (dissociation curve). Primers had efficiency values between 80% and 110%, and each pair amplified a single product in dissociation curve analyses. Amplicons were purified using SureClean (BioLine, Taunton, Massachusetts, USA) and sequenced as previously reported (
Sutherland et al. 2011). Samples were diluted 20-fold in water, and each was run in duplicate on a single plate. Replicates were within 0.5 Ct for 193 of 195 sample-target combinations. Each gene/plate included a no RT control and an NTC. Negative controls did not amplify.
Data were exported from MxPro software (Agilent), imported into R, and analyzed using ReadqPCR and NormqPCR (
Perkins et al. 2012;
R Core Team 2017). Potential normalizers
vcl,
gstd2,
flna, and
rps20 were evaluated for stability using geNORM (
Vandesompele et al. 2002) implemented within NormqPCR; the geometric mean of the two normalizers with the lowest
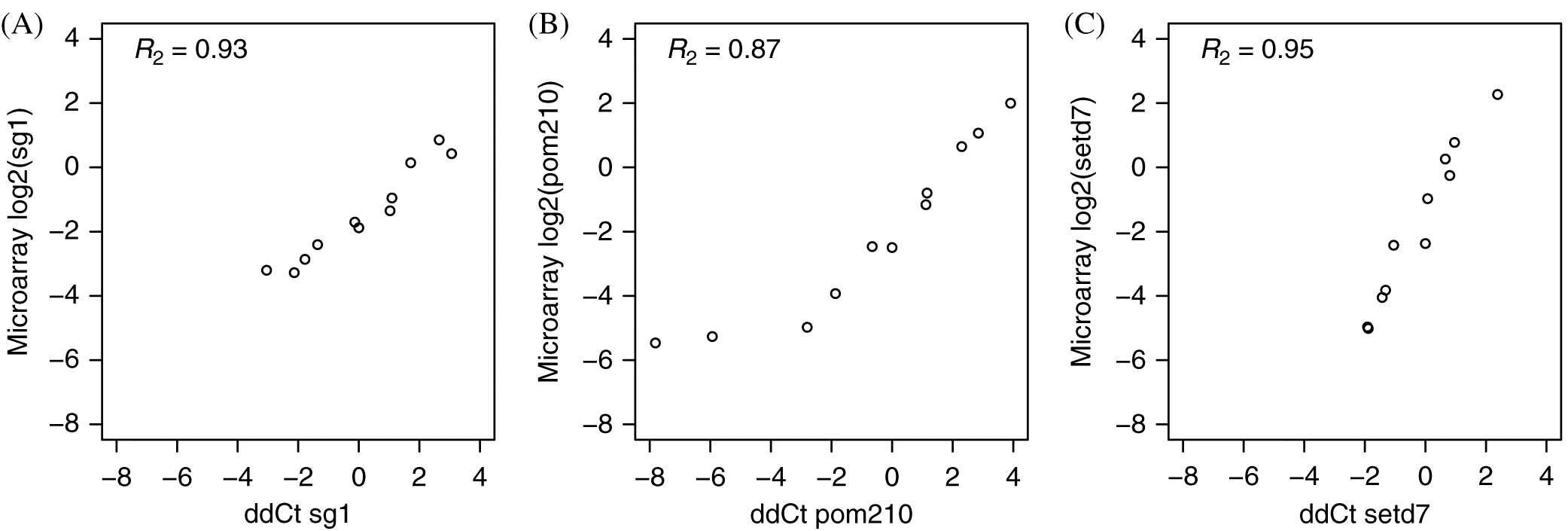
M-value was used to normalize samples. Delta-delta Ct (ddCt) values were calculated by first subtracting the geometric mean of the normalizer genes (dCt within NormqPCR) and then subtracting this value from the median dCt values for this gene. These ddCt values (i.e., log
2) were used to correlate against the log
2 microarray ratios to obtain the adjusted
R2 value from the linear model fit in R (
R Core Team 2017).
Discussion
As drug resistance continues to evolve against parasiticides used in salmon aquaculture, it will be important to continue considering complementary methods of parasite control, such as host immunostimulation. The viability of this option requires an effective removal of lice outweighing any negative effects on hosts. This can be intermittently used to boost the host immune response; for example, when no vaccine is available against the pathogen (
Casadei et al. 2013). No commercially available vaccine exists for salmon lice
L. salmonis, although efforts towards this goal have been made (
Raynard et al. 2002).
Targeting the induction of appropriate defenses and immune system components for specific pathogens is important when considering defense (
Medzhitov 2007). In rainbow trout, dietary peptidoglycan is being explored as a potential treatment to increase the expression of antimicrobial peptides for elevating immune responses to common bacterial diseases (
Casadei et al. 2013). As our knowledge on the appropriate responses to ectoparasites improves (in this case against salmon lice), immunostimulation formulations can be optimized towards the specific parasite (
Alvarez-Pellitero 2008).
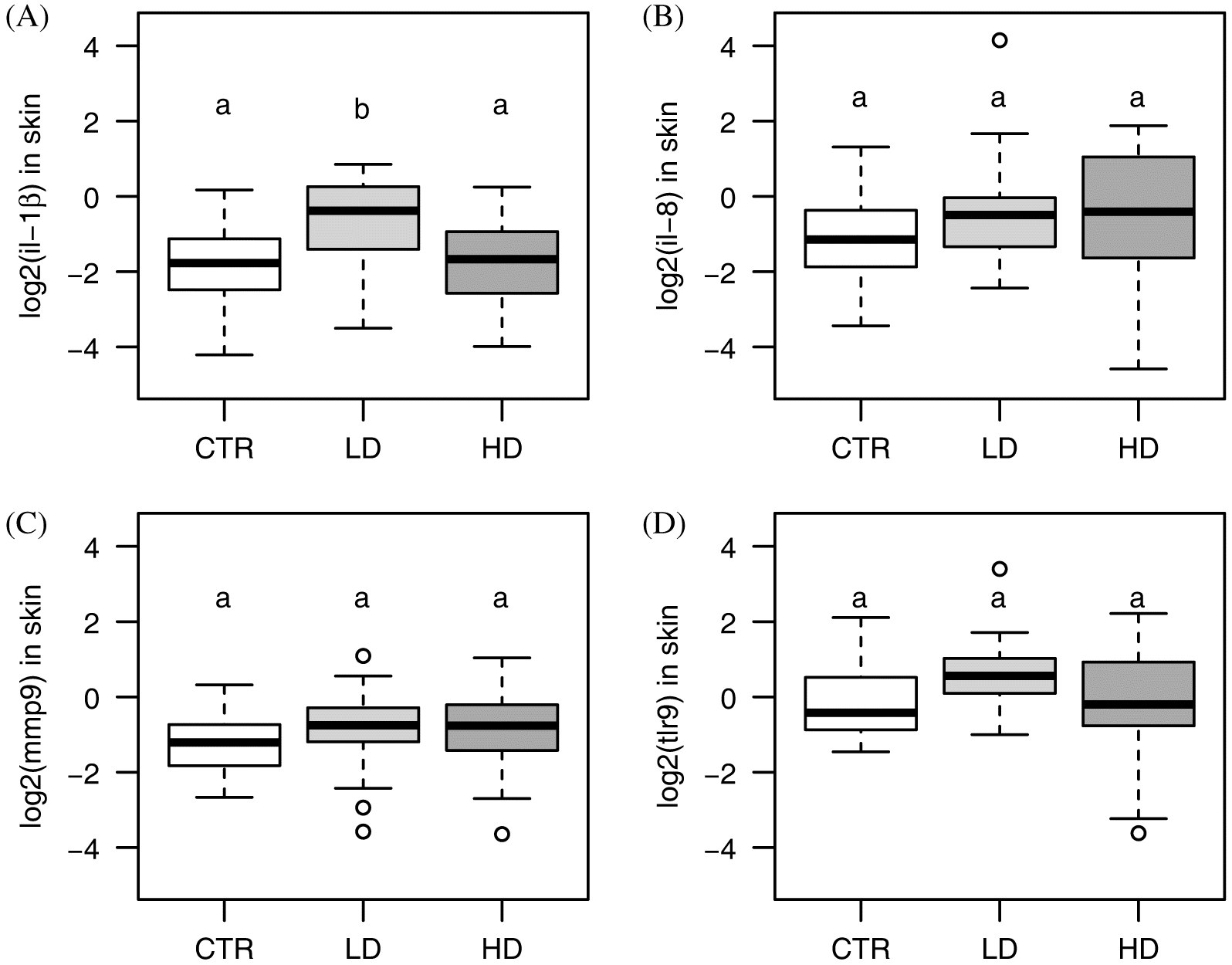
Pro-inflammatory responses have long been understood to play an important role in salmon lice rejection (
Fast 2014). Increased
il-1β expression and innate defenses are important for host rejection of salmon lice; this response is not typical of Atlantic salmon, but rather is typical of the more lice-resistant pink salmon and coho salmon (
Johnson and Albright 1992;
Jones et al. 2007;
Braden et al. 2012,
2015;
Sutherland et al. 2014). However, in previous studies of CpG-ODN immunostimulation, Atlantic salmon fed the treatment diet overexpressed
il-1β and had lower lice infection levels than controls (
Covello et al. 2012;
Poley et al. 2013). Similarly, here,
il-1β was overexpressed in the LD salmon, the group with the lowest infection levels. Although
tlr9 was overexpressed in the group fed the immunostimulant diet, the expression of this gene decreased over time. Atlantic salmon CpG-ODN has been shown to bind
tlr9 for signaling IFN-related genes (
Iliev et al. 2013); it is possible that the current immunostimulant is working through the same pathway, but this would require more work to confirm. Surprisingly,
mmp9 was not increased in mRNA levels from the infection, as in many other studies, this gene is a reliable indicator of a response to lice infection. The maintenance of a low-level lice infection in this study may not have been sufficient to trigger this response. The biphasic nature of the response of
mmp9 has been previously identified (
Tadiso et al. 2011) and can change throughout the infection. Although a larger panel of response genes or a full transcriptome profiling would be needed to better understand the host response to the immunostimulant, the expression of
il-1β coincident with the lower infection levels suggests that the LD immunostimulant enhanced inflammation, directing the Atlantic salmon response towards a more successful lice rejection response.
The overexpression of
il-1β in LD salmon was specific to the skin (the point of
L. salmonis contact), with minor overexpression (∼1.4-fold) also observed in the spleen, a secondary hematopoietic organ for salmon. Mucosal immunity is important for ectoparasite defense, and the highest expression changes may be observed in the tissue contacting the ectoparasite (for review, see
Alvarez-Pellitero 2008;
Braden et al. 2012). Notably, the transcriptome response of the skin of immunostimulated Atlantic salmon infected with
Caligus rogercresseyi provided more information on the host defense response than the expression analysis of the host head kidney (
Núñez-Acuña et al. 2015). In the present study,
il-1β levels decreased over time in the anterior kidney (the primary hematopoietic organ of fish;
Supplementary Material 2). It is possible that this reflects a shifting tissue distribution of cells expressing this gene, moving from the source to the site of external contact. Further work would be required to confirm this tissue change. Up-regulation of
il-1β in the skin in response to
L. salmonis has been previously observed. For example,
il-1β expression was up-regulated specifically in the skin of infected pink salmon (considered to be more resistant) but not Atlantic salmon (
Sutherland et al. 2014). Considering the potential for immunosuppression of Atlantic salmon by lice-derived PGE
2 and other compounds (
Fast et al. 2004,
2007), it is interesting that the immunostimulation induced a response more characteristic of a successful rejection, as immunostimulation can negate immune suppression (for review see
Vadstein 1997). Up-regulation of
il-1β promotes a T helper 17 response that is important in defense against salmon lice (
Skugor et al. 2008), bacteria, and fungi, but is also implicated in auto-immunity (
Waite and Skokos 2012), highlighting the importance of dose specificity in levels of effector induction. The response of the LD salmon more reflects the response of pink salmon, suggesting a shift towards a more appropriate defense mechanism.
Although not significant (
p = 0.07), there was a trend towards the overexpression of
il-8 in the LD salmon; an insignificant trend towards overexpression was also previously observed in Atlantic and pink salmon skin (
Sutherland et al. 2014). Earlier work found elevated
il-8 expression in head kidney/spleen pooled tissues at 7 and 14 d post infection in juvenile chum (
Oncorhynchus keta) and pink salmon but no consistent response in
il-1β (
Jones et al. 2008a). Similarly,
il-8 was found to be up-regulated at 7 d post infection in pink salmon anterior kidney and skin (
Jones et al. 2007). In the present study, up-regulation of
il-1β occurred consistently in LD-fed Atlantic salmon (the feed group with the lowest infection), as it did in pink salmon skin in previous studies, also coinciding with early responses (6 d after exposure;
Sutherland et al. 2014). Between HD and LD, the LD was optimal here in terms of
il-1β induction and reduction in lice infection levels.
Genes underexpressed in lice parasitizing LD salmon included protein degradation genes potentially involved in feeding on susceptible hosts such as Atlantic salmon (e.g., metalloproteinases, chymotrypsin, and trypsin-like serine protease;
Braden et al. 2017). Lower activity and presence of similar enzymes was previously observed when lice were exposed to mucus from less optimal hosts (e.g., coho salmon), unsuitable hosts (e.g., winter flounder,
Pseudopleuronectes americanus), or seawater (
Fast et al. 2003). Three additional genes underexpressed in LD-feeding lice included the protease inhibitor
papilin and two genes annotated to
L-amino-acid oxidase, an enzyme found in snake venom (
Du and Clemetson 2002). The
papilin sequence contains a protease inhibitor domain (kunitz; CDD: pfam00014) frequently observed in tick salivary secretions (
Francischetti et al. 2009;
Tirloni et al. 2014), though the oxidases could have roles in virulence based on their annotations to the Malayan pit viper (
Calloselasma rhodostoma) and Australian taipan (
Oxyuranus scutellatus). More work is needed to understand exactly the relation of specific transcripts to the feeding response of
L. salmonis; however, this suggests that the feeding response of lice infecting LD salmon may be reduced.
An analysis of immunostimulation on the Caligid sea lice (
Caligus rogercresseyi) identified four ionotropic receptors as differentially expressed in Atlantic salmon exposed to 1% peptidoglycan in-feed (
Núñez-Acuña et al. 2016). A
tblastx search (e < 10
−10) of the
L. salmonis transcriptome using these
C. rogercresseyi sequences as a query resulted in the identification of their putative orthologues. The four
C. rogercresseyi genes
ionotropic kainite 2 receptor (
KAR-2; GenBank: KJ002538.1),
KAR-2-like-a (KJ002539.1),
KAR-2-like-b (KJ002540.1), and
ionotropic glutamate 25a receptor (KJ002537.1) matched
L. salmonis probes C045R029, C139R121, C082R071, and C217R137, respectively. Although these probes passed quality filters (Supplementary Material 3), they were not differentially expressed here.
Genes overexpressed in lice parasitizing LD salmon were those related to lice survival on immunostimulated hosts and may provide insight into mechanisms of host immune evasion. Interestingly, carboxylesterases were abundant in this list of response genes. It will be important to continue exploring genes that may provide protection against the innate immunity of various salmonids. Interestingly, immune-related genes in the lice were also slightly overexpressed when feeding on immunostimulated fish, including
granulin-7,
saposin-D, and
B-cell receptor-associated protein 31. It remains unclear if the parasite consumes any of the immunostimulant through feeding on host mucous, skin, or blood during infection, and if this consumption can impact its physiology. As carboxylesterases also have roles in immunity in arthropods (
Johansson et al. 2005;
Albert et al. 2011), it will be important to determine if lice can be immunostimulated through feeding on treated hosts, and how this might impact louse responses to other treatments. However, determining whether the louse response is due to an enhanced host response or directly to the immunostimulant is not possible here.
Some similarities in identities of differentially expressed lice genes were present between lice-infecting LD salmon and lice with elevated resistance to EMB (trade name: SLICE). For example, putative collagenases were underexpressed in LD-feeding lice but overexpressed in EMB-resistant lice (
Sutherland et al. 2015), such as
trypsin-like serine protease, and
nas-6 and
-14. Also, collagenases were overexpressed in EMB-resistant males (i.e., the most resistant group of
L. salmonis in
Sutherland et al. 2015), and here, lice-infecting LD salmon overexpressed
setd7, a histone methyltransferase chromatin remodeler involved in induction of collagenase transcription (
Martens et al. 2003). Interestingly, a link between immunostimulation and EMB response has been previously identified in
L. salmonis (Atlantic subspecies); lice feeding on CpG-ODN immunostimulated hosts showed higher EMB tolerance than those on normal hosts (
Poley et al. 2013). The authors propose that genes such as monooxygenases could be induced by feeding on an immunostimulated host, and that this increased expression of monooxygenases could prime lice against EMB or buffer against consequences of EMB exposure. In another study, lice feeding on immunostimulated hosts had elevated
p-glycoprotein expression, a potential resistance mechanism against EMB (
Igboeli et al. 2012), and had higher survival after infecting EMB-fed salmon (
Igboeli et al. 2013). It will be important to determine effective combinations of control strategies, as they may not all be compatible. More work would be required to understand the molecular signaling involved in these responses and the potential antagonistic or synergistic effects of prior host immunostimulation and subsequent chemical interventions.