Shining light on recent advances in microbial mercury cycling
Abstract
Mercury (Hg) is a global pollutant emitted primarily as gaseous Hg0 that is deposited in aquatic and terrestrial ecosystems following its oxidation to HgII. From that point, microbes play a key role in determining Hg’s fate in the environment by participating in sequestration, oxidation, reduction, and methylation reactions. A wide diversity of chemotrophic and phototrophic microbes occupying oxic and anoxic habitats are known to participate directly in Hg cycling. Over the last few years, new findings have come to light that have greatly improved our mechanistic understanding of microbe-mediated Hg cycling pathways in the environment. In this review, we summarize recent advances in microbially mediated Hg cycling and take the opportunity to compare the relatively well-studied chemotrophic pathways to poorly understood phototrophic pathways. We present how the use of genomic and analytical tools can be used to understand Hg transformations and the physiological context of recently discovered cometabolic Hg transformations supported in anaerobes and phototrophs. Finally, we propose a conceptual framework that emphasizes the role that phototrophs play in environmental Hg redox cycling and the importance of better characterizing such pathways in the face of the environmental changes currently underway.
Introduction
Mercury (Hg) is a global pollutant emitted from natural sources such as volcanic activity and forest fires and anthropogenic sources such as mining and coal combustion (Obrist et al. 2018). Hg is emitted primarily in its elemental and highly volatile form Hg0, which can travel in the atmosphere for up to a year before being deposited in aquatic and terrestrial ecosystems as oxidized HgII (Selin et al. 2007). From that point, Hg can be subject to a variety of transformations including burial into sediments; reduction, which results in Hg0 re-emission to the atmosphere; and methylation, which converts inorganic Hg to toxic monomethylmercury (MeHg) that bioaccumulates in animals (Sunderland 2007) and plants such as rice (Zhang et al. 2010).
Microbes play a key role in Hg transformations. They can produce or degrade MeHg, reduce HgII, oxidize Hg0, and sequester a variety of Hg chemical species (as summarized by Barkay and Wagner-Dobler 2005 and Grégoire and Poulain 2014). Generally speaking, microbial Hg transformations occur through cometabolic processes (e.g., encoded by the hgcAB gene cluster) or dedicated detoxification strategies (e.g., encoded by the mer operon). Hg sequestration can occur through dedicated pathways (e.g., phytochelatins binding Hg (Rauser 1990; Kawakami et al. 2006)) or cometabolic processes (e.g., HgS formation during photosynthesis (Kelly et al. 2007)). Indeed, a wide diversity of microbes can mediate Hg cycling pathways in a variety of habitats; aerobic, anaerobic, chemotrophic, and phototrophic microbes all influence the fate of Hg in the environment.
In an earlier review, we synthesized the state of knowledge on Hg transformations mediated by phototrophic microbes (Grégoire and Poulain 2014). We highlighted that phototrophic Hg cycling pathways were poorly understood compared with their chemotrophic counterparts. We also discussed why this was an important knowledge gap to address because of the drastic changes predicted to occur in phototrophic communities in response to global environmental changes such as the increased frequency, magnitude, and duration of phytoplankton blooms (Krabbenhoft and Sunderland 2013).
Although the environmental contributions of phototrophs to environmental Hg cycling remain understudied compared with their chemotrophic counterparts, considerable research has been done in recent years to better understand the role of phototrophs in the global Hg cycle. Here, we offer an update to our initial literature review comparing chemotrophic and phototrophic Hg cycling pathways including 68 research papers published since 2014 (Grégoire and Poulain 2014). Our objective with this review was to summarize the recent advances in our understanding of chemotrophic and phototrophic Hg cycling pathways. As part of this, we highlight outstanding knowledge gaps pertaining to microbially mediated Hg cycling emphasizing recent efforts that have helped better understand the role of phototrophs in Hg cycling. Ultimately, we aim to provide recommendations on how to better characterize the environmental and ecological context of microbial Hg cycling in future research.
Chemotrophic mercury transformations
Chemotrophic mercury methylation
Chemotrophic Hg methylation continues to be the most studied microbially mediated Hg transformation, which is unsurprising given the health concerns surrounding MeHg. At the time of our initial review, the discovery of the hgcAB gene cluster encoding for Hg methylation had just been published (Parks et al. 2013). Since this transformational finding in the field of Hg research, there has been a surge in the number of studies investigating the physiological and environmental controls of Hg methylation.
On the mechanistic front, computer modeling has shown that the corrinoid protein encoded by hgcA initiates a key step in MeHg formation by transferring a negatively charged methyl carbanion to an HgII substrate (Zhou et al. 2014). This process is thought to occur on the inner side of the cell membrane based on observations in spheroplasts (i.e., bacteria partially/wholly devoid of a cell wall) generated from well-characterized Hg-methylating bacteria (Schaefer et al. 2014b). With respect to the physiology of Hg methylation, gene deletions targeting hgcAB in Geobacter sulfurreducens PCA, a well-known Hg methylator with proven Hg redox cycling capabilities, resulted in increased Hg uptake, HgII reduction, and Hg0 oxidation (Lin et al. 2014a; Qian et al. 2016). In contrast, deleting the genes encoding for the electron transport machinery essential to HgII reduction (e.g., encoding for cytochrome c synthesis) in G. sulfurreducens increased the abundance of proteins involved in the metabolism of C1 compounds potentially involved in Hg methylation (Qian et al. 2016). These studies suggest that Hg methylation and Hg reduction can compete for common Hg or metabolic substrates inside the cell and support the presence of common physiological controls for both transformations. Further physiological characterization of strains capable of a suite of Hg transformations are required to identify potential the metabolic coupling points that exist for competing Hg transformations.
The discovery of the hgcAB gene cluster has revealed a broader diversity of microbes that can potentially methylate Hg compared with what was known four years ago (Podar et al. 2015). Previously, representatives within the sulphate-reducing bacteria (SRB) (Choi et al. 1994a, 1994b), iron-reducing bacteria (Fleming et al. 2006) and methanogens (Han et al. 2010; Hamelin et al. 2011; Yu et al. 2013) were known to methylate Hg. Homologues for hgcAB have now been identified in representatives of the phyla Chloroflexi and Firmicutes (Schaefer et al. 2014a; Christensen et al. 2016); a compilation of microbial strains harbouring the hgcAB sequence has been published elsewhere (Paranjape and Hall 2017). Most recently, robust Hg methylation was demonstrated in several cultured methanogen strains (Gilmour et al. 2018). Such studies exemplify how hgcAB genes can be used to identify Hg methylators across diverse clades of microbes and provide insight into the environmental controls of Hg methylation that exist in a variety of habitats.
Mining existing (meta)genomic information for hgcAB sequences has revealed several new environments that can potentially support Hg methylation including invertebrate digestive tracts, permafrost soil, hypoxic coastal areas, soda lakes, and thermal springs (Podar et al. 2015). In addition, a number of field studies have evaluated the presence or quantified the abundance of hgcA gene or hgcAB genes in habitats including wetland sediments (Bae et al. 2014; Schaefer et al. 2014a; Graham et al. 2018), wastewater treatment plants (Bravo et al. 2018), rivers near chlor-alkali plants (Bravo et al. 2016), lake sediments (Ma et al. 2017b), river biofilms (Dranguet et al. 2017), hydroelectric dam sediments (Ma et al. 2017a), rice paddies (Su et al. 2016; Vishnivetskaya et al. 2018), and Antarctic sea ice (Gionfriddo et al. 2016). If the number of studies cited above is any indication, the number of habitats that can support Hg methylation will continue to grow, which will help identify novel and potentially overlooked Hg methylation hotspots in the environment.
Several of the above-mentioned studies have assessed the distribution the hgcAB gene cluster alongside the abundance of marker genes used as proxies for microbial metabolisms that support Hg methylation. Studies using genes such as the dsrAB genes (encoding for subunits of the dissimilatory sulphite reductase (EC 1.8.99.5)) and the mcrA gene (encoding the methyl-coenzyme M reductase (EC: 3.1.21)) as proxies for sulphate reduction and methane cycling, respectively, have linked the abundance of Hg methylation genes to different metabolic functional groups (Bae et al. 2014; Gionfriddo et al. 2016; Dranguet et al. 2017; Ma et al. 2017b; Bravo et al. 2018). These studies have led to the discovery that syntrophic microbes, such as those that rely on interspecies hydrogen and acetate transfer following propionate fermentation for growth, play an important role in Hg methylation (Bae et al. 2014; Bravo et al. 2018) in environments depleted in sulphate and iron (e.g., terminal electron acceptors) (Yu et al. 2018). The findings concerning syntrophs highlight the importance of moving away from studies that use single model organisms to better address how complex microbial assemblages can influence MeHg production in the environment.
Despite the surge in studies investigating Hg methylation in microbes, whether a physiological purpose for this Hg transformation exists remains unclear. Hg methylation was once thought to be a detoxification strategy, as studied in the fungus Neurospora crassa (Landner 1971). However, experimental evidence in other microbes, including bacteria and archaea, are currently lacking. Much of the evidence published to date supports that Hg methylation is a cometabolic, accidental process. In support of cometabolic Hg methylation, recent studies have shown that organic carbon source composition (Bravo et al. 2016; Christensen et al. 2018), electron acceptor availability (Bravo et al. 2018; Yu et al. 2018), and salinity (Bravo et al. 2016) appear to control the distribution hgcAB genes. With some of the genetic determinants for Hg methylation now in hand, the field is ripe to explore the physiological and evolutionary context of Hg methylation in a variety of model organisms.
Despite the recent emphasis on the genetic basis of Hg methylation, research addressing the fundamental aspects of Hg bioavailability to methylators continues to expand. Previously, it was thought that Hg methylation was limited by the availability of inorganic Hg substrates such as HgS, which was considered poorly bioavailable to methylators in sulphidogenic environments (Liu et al. 2009). The importance of HgS as a substrate for methylation has recently been revisited in the context of how HgS nanoparticle formation controls Hg bioavailability to methylators under anoxic conditions. It has been shown that HgS particles tend to agglomerate, becoming more crystalline and less bioavailable as they age in the presence of dissolved organic matter (DOM), which can decrease Hg methylation (Pham et al. 2014). In contrast, it has also been shown that methylation of HgS can increase in the presence of DOM as a function of thiol content, preventing large and poorly bioavailable HgS aggregates from forming (Graham et al. 2013; Graham et al. 2017). Aside from thiols, it has been suggested that amine and, to a lesser degree, carboxyl functional groups, can also control HgS aggregation once the binding sites of thiols have been saturated at high Hg to DOM ratios (Mazrui et al. 2018). These studies suggest that the controls on HgS nanoparticle formation are complex and reveal the transient nature of how HgS species exert dynamic controls on Hg bioavailability to Hg methylators.
The bioavailability and toxicity of Hg are controlled, in part, by its affinity for thiol-bearing molecules such as cysteine. For the sake of brevity, we have omitted discussing mechanisms for Hg uptake and toxicity in this section as these topics have been recently reviewed elsewhere (Hsu-Kim et al. 2013; Grégoire and Poulain 2014; Parks and Smith 2016; Mahbub et al. 2017). Instead, we briefly focus on recent work addressing the role of DOM and thiol-bearing ligands in mediating Hg bioavailability to Hg methylators.
In a recent study using the model Hg-methylating bacterium G. sulfurreducens PCA, cysteine inhibited Hg methylation at low concentrations (0.01 to 0.1 μM) but stimulated Hg methylation at higher concentrations (100 to 1000 μM) when experiments were run for 144 h vs 4 h (Lin et al. 2015). One potential explanation provided for this trend was that thiols mobilized Hg from binding sites on the cell membrane to the cytoplasm, where Hg was more bioavailable for methylation (Liu et al. 2016). The role of functional groups in the cell membrane was recently highlighted in a study comparing Hg methylation in Desulfovibrio desulfuricans ND132 and G. sulfurreducens in the presence of two sources of DOM (one of aquatic origin with low aromaticity and the other of terrestrial origin with relatively higher aromaticity) (Zhao et al. 2017). In this study, MeHg production by D. desulfuricans increased with DOM concentration, whereas MeHg production in G. sulfurreducens decreased (Zhao et al. 2017). Another study examining Hg stable isotope fractionation during Hg methylation in D. desulfuricans and G. sulfurreducens demonstrated that the same strains accessed different intracellular and extracellular pools of Hg during methylation (Janssen et al. 2016). These studies highlight that strain-specific characteristics can have a considerable impact on Hg uptake and subsequent methylation. Although the exact nature of the differences in characteristics has yet to be properly defined, they are clearly important to consider when extending conclusions drawn from a given model Hg methylator to microbial communities in the environment.
Chemotrophic methylmercury demethylation
Compared with Hg methylation, MeHg demethylation has been relatively understudied, although new mechanistic details have emerged for both reductive demethylation (RD) and oxidative demethylation (OD) pathways. Microbes capable of RD harbour the mer operon, which is a series of genes encoding dedicated Hg scavenging, transport, and detoxification machinery (Barkay et al. 2003; Parks et al. 2009). The mer operon’s expression is induced as a function of intracellular Hg concentration and RD is carried out thanks to the presence of merB, which encodes an organomercury lyase (e.g., MerB) that cleaves MeHg into inorganic HgII and CH4 (Parks et al. 2009). HgII is subsequently reduced to Hg0 via the mercuric reductase MerA and CH4 and Hg0 can evade the cell (Barkay et al. 2003). In contrast with RD, OD occurs in the absence of dedicated Hg detoxification machinery and is a non-specific cometabolic process tied to C1 compound metabolism that results in the production of HgII and CO2 (Oremland et al. 1991; Hsu-Kim et al. 2013).
Recent work on MerB has shown that conserved cysteine and aspartic acid residues are essential for releasing Hg from MerB’s active site following the cleaving of the methyl group (Silva and Rodrigues 2015). When a serine residue was present in place of aspartic acid, MerB lost its specificity for MeHg and was also able to bind copper (Cu), although resupplying MerB with Hg successfully removed Cu from the enzyme’s active site (Wahba et al. 2016). These findings demonstrate that MeHg can outcompete Cu for the binding site on MerB, but also highlight functional parallels that may exist between RD and Cu homeostasis that merit further investigation.
Despite MeHg formation occurring predominantly in anoxic environments, broad Hg resistance strategies relying on MerB have largely been associated with aerobes. Recent work with the model anaerobe Geobacter bemidjiensis Bem suggests that strategies similar to those encoded by the mer operon may be present in anaerobes providing them with a means for RD (Lu et al. 2016). Geobacter bemidjiensis supported a suite of Hg transformation pathways including Hg methylation, HgII reduction via a MerA-like mechanism, and Hg0 oxidation (Lu et al. 2016). The latter finding is interesting because, to the best of our knowledge, very few complete mer operons have been reported for obligate anaerobes. Although these are not the first observations that anaerobes can mediate several Hg transformations, they do provide an interesting basis from which to investigate the physiological controls of competing Hg transformations that may occur simultaneously.
Similar to the work done on Hg methylation, new studies have addressed how thiol-bearing ligands affect RD. In contrast with inorganic Hg, thiol-bearing ligands had little effect on MeHg bioavailability in bacteria capable of RD (Escherichia coli and Pseudomonas stutzeri) (Ndu et al. 2016). The same study found a considerable difference in MeHg demethylation rates for the two strains despite comparable bioavailability among the different MeHg complexes tested (Ndu et al. 2016). Similar to what was suggested for Hg methylators, the authors attributed the contrasting results to differences in MeHg bioavailability associated with strain specific characteristics (Ndu et al. 2016). Although the authors demonstrated that non-specific binding of MeHg to heat-killed cells rendered a portion of the MeHg unavailable for demethylation, the exact nature of these binding sites was not further discussed (Ndu et al. 2016)
A newly discovered OD pathway in methanotrophs serendipitously provides additional links between Cu metabolism and MeHg demethylation. Methanobactin, a Cu chaperone molecule synthesized by methanotrophs, was essential for OD (Baral et al. 2014; Lu et al. 2017). Recent findings in the model methanotroph Methylosinus trichosporium OB3b showed that MeHg demethylation was dependent on the activity of methanol dehydrogenase (Lu et al. 2017), further supporting a link between C1 metabolism and OD (Hsu-Kim et al. 2013). Given that C1 metabolism is also tied to Hg methylation, these results suggest that the availability of C1 compounds in the environment plays an important role in mediating MeHg accumulation. That being said, when considering the role of methanotrophs in OD and its relevance to the environment, one must remember that in most environments where methanotrophs are active, MeHg degradation via OD could be overwhelmed by the large availability of other reduced C1 compounds (e.g., methane). The environmental relevance of this type of OD remains to be determined.
At the environmental scale, a number of studies have measured MeHg demethylation (many of which were summarized by Paranjape and Hall 2017); however, few of these studies explicitly discuss the direct contribution of microbes to demethylation. The fact that many of these systems are net MeHg sources makes it difficult to investigate the mechanisms supporting MeHg destruction beyond the laboratory scale. Further investigations into the mechanisms of biotic MeHg sinks may yield important clues to mitigating MeHg exposure in the environment. To the best of our knowledge, only one recent study has been published that fits this description. In this work, Kronberg et al. 2018 showed that methanogens are important contributors to OD in a wetland ecosystem known to be a sink for MeHg. Together with previous work on methanogens, this finding frames methane cycling microbes as key players in controlling MeHg levels in the environment.
Chemotrophic mercury reduction
The mercuric reductase MerA encoded by the mer operon continues to be one of the best-studied chemotrophic Hg reduction strategies. Recently, it has been shown that sulphur-oxidizing bacteria carrying merA were not only able to reduce HgS minerals in aquatic environments, but also use HgS as a sulphur source (Vazquez-Rodriguez et al. 2015). Although it is well established that MerA relies on the use of nicotinamide adenine dinucleotide phosphate (NADPH) as a redox cofactor for HgII reduction (Barkay et al. 2003), the link between mer-mediated reduction and the redox state of the cell is rarely discussed. Recent work with Thermus thermophilus HB27 revealed important links between cellular redox balance, Hg sequestration and MerA activity (Norambuena et al. 2018). Thermus thermophilus (alongside other representatives of the Thermus genus) harboured genes upstream of merA in the mer operon encoding for the synthesis of low molecular weight thiols that buffered Hg toxicity prior to HgII reduction (Norambuena et al. 2018). The synthesis of cellular redox buffering compounds proved essential to supporting this Hg detoxification strategy (Norambuena et al. 2018).
Perhaps by further investigating the influence of redox homeostasis on mer-mediated reduction we can gain insights into how such Hg detoxification strategies evolved across different redox conditions throughout Earth’s history (Barkay et al. 2010). Therein may lay an explanation for the observation of a MerA-like reductase in the obligate anaerobe G. bemidjiensis despite such strategies being largely absent in anaerobes (see the previous section). Perhaps such pathways only occur under specific redox conditions in the environment that support anaerobic metabolism optimized for cell growth and Hg detoxification.
Anaerobes such as dissimilatory metal-reducing bacteria continue to be studied with respect to non-mer-mediated Hg reduction pathways. As previously mentioned, work with G. sulfurreducens has shown that HgII reduction increased following the deletion of hgcAB (Lin et al. 2014a), as did the abundance of cytochrome-c proteins supporting HgII reduction (Qian et al. 2016), supporting a physiological link for both of these Hg transformations. Recently, it has also been shown that HgII reduction in Shewanella oneidensis MR-1 increased in the presence of natural organic matter (NOM) (Lee et al. 2018). In this instance, the authors indicated that HgII reduction increased as a function of electron transfer from S. oneidensis to NOM, which led to the formation of free radicals that could reduce HgII (Lee et al. 2018).
Novel non-mer-mediated pathways have also recently been discovered in previously untested organisms. Magnetotactic bacteria are now known to reduce HgII during microaerophilic growth via coupled redox reactions with biogenic magnetite in the membrane of the magnetosome (Liu and Wiatrowski 2018). Our own research has revealed that HgII reduction is supported in obligate anaerobes from the order Clostridiales. We observed that HgII reduction was dependent on the ability of cells to produce reduced redox cofactors (possibly ferredoxin) via fermentation when pyruvate was used as a carbon source (Grégoire et al. 2018). Given that merA was absent in these strains, our results support that cells reduced Hg because of its electrophilic nature allowing cells to transfer electron from reduced redox cofactors to Hg during fermentative growth (Grégoire et al. 2018). This finding suggests that fermentative Hg reduction may be an important pathway for Hg redox cycling in environments devoid of light and electron acceptors (Grégoire et al. 2018). Given that fermentative microbes occupy a niche similar to that of chemotrophic Hg methylators (Desrochers et al. 2015), such pathways may also influence the accumulation of MeHg. That being said, the contributions of fermenters to HgII reduction and the potential influence of such pathways on Hg availability to methylators have yet to be investigated.
Chemotrophic mercury oxidation
Hg oxidation still remains one of the most poorly understood chemotrophic Hg transformations, with the first mechanistic details having only just recently been published. Following the seminal work demonstrating Hg0 oxidation via redox reactions with thiols in the cell membranes of anaerobes (Colombo et al. 2013), additional evidence has emerged supporting the presence of a similar pathway in other obligate and facultative anaerobes (Colombo et al. 2014; Lin et al. 2014b; Lu et al. 2016). It has also been shown that dead cell material bearing intact thiol groups can catalyze Hg0 oxidation offering a potentially important route for delivering freshly oxidized HgII to anoxic environments (Colombo et al. 2014). Hg0 oxidation is thought to be more important in situations where thiol binding site availability is high relative to low extracellular Hg concentration (Lin et al. 2014b). Given the ubiquitous nature of thiols in bacterial membranes (Yu et al. 2014), Hg0 oxidation by anaerobes is likely widespread in environments such as waterlogged soils (Mazur et al. 2015; Poulin et al. 2016) and anoxic lake sediments (Bouffard and Amyot 2009) where Hg0 can dominate Hg speciation. When considered alongside the growing evidence for anaerobic HgII reduction, these findings show that anaerobes are poised to be key players in controlling MeHg production in anoxic systems by catalyzing a dynamic anaerobic redox cycle.
Phototrophic microbial mercury transformations
As outlined in our previous review, there is a growing body of evidence suggesting that oxygenic and anoxygenic phototrophs can participate in Hg transformations in the environment (Grégoire and Poulain 2014). Phototroph-mediated Hg cycling pathways stand to be important in environments where abiotic photochemical Hg transformations are limited (Poulain et al. 2004). Such habitats could include the metalimnion of thermally stratified lakes (Poulain et al. 2004), microbial mats (Dupraz and Visscher 2005; Polerecky et al. 2007), and photic surface sediments (Sloth et al. 1996; Rossi et al. 2012) where phototrophs thrive at low light intensities. Despite the potential for phototrophs to directly influence Hg speciation, considerably fewer studies have addressed phototrophic Hg cycling in recent years compared with the large body of work that exists on chemotrophs. That being said, recent research at the laboratory scale has advanced our mechanistic understanding of Hg uptake, toxicity, and transformation in a variety of phototrophic organisms.
Phototrophic mercury uptake studies
In our initial review, we detailed the existing body of work on the passive and active Hg uptake pathways in phototrophs (Grégoire and Poulain 2014). At that time, all of the information available on Hg uptake in phototrophs was derived from oxygenic phototrophs such as algae, cyanobacteria, and diatoms. Since then, new mechanistic details have emerged on the uptake of Hg in the model green alga Chlamydomonas reinhardtii and, for the first time, in anoxygenic phototrophic purple non-sulphur bacteria (PNSB).
Transcriptomic evidence has suggested that C. reinhardtii can actively take up inorganic Hg complexes through divalent metal transporters, and organic Hg complexes through amino acid transporters (Beauvais-Fluck et al. 2017), although this was not directly experimentally tested. A similar mechanism was proposed for model PNSB whereby Hg uptake occurred via Ca2+ transport channels as an active process (Kis et al. 2017). These findings echo those for chemotrophic anaerobes where active uptake of Hg occurred through Zn2+ or Mn2+ transporters (Schaefer et al. 2014b; Stenzler et al. 2017). These studies suggest that Hg uptake via divalent cation transporters is supported across diverse microbial metabolisms. Given the high metal requirements associated with phototrophy, Hg uptake via metal transporters is possibly important and may exert a strong influence on downstream Hg transformations.
Whereas the effects of DOM on Hg bioavailability to chemotrophs have been extensively studied, there is little information available for phototrophs (Zhong and Wang 2009). Microbial phototrophy is not limited to autotrophy. Strategies such as mixotrophy where cells can use light for energy and a diverse carbon pool for biosynthesis, could lead to Hg uptake via the shuttling of Hg-DOM complexes into cells (Chiasson-Gould et al. 2014). To the best of our knowledge, the contributions of mixotrophs to Hg uptake in aquatic food webs have only recently been addressed (Cárdenas et al. 2014; Soto Cárdenas et al. 2018). In these studies, the authors showed that mixotrophic ciliates could incorporate Hg through passive diffusion and active uptake linked to bacterivory (Cárdenas et al. 2014; Soto Cárdenas et al. 2018). Despite mixotrophy being widespread in the environment (Raven 2009), the potential implications of mixotrophy on Hg accumulation in aquatic and terrestrial food webs remain largely unexplored.
Phototrophic mercury methylation
The majority of studies addressing the role of phototrophs in controlling MeHg’s fate focus on bioaccumulation in food webs, and fewer studies directly address how phototrophic communities affect Hg speciation in the environment. Most of the recent work has been conducted on periphytic communities. These studies face the challenge of distinguishing the direct involvement of phototrophs in Hg methylation from the indirect role that they have (as primary producers) in supplying nutrients to chemotrophic Hg methylators in the periphytic matrix (Lanza et al. 2017; Bouchet et al. 2018). In most cases, MeHg production increased under conditions of higher photosynthetic activity (Gionfriddo et al. 2016; Olsen et al. 2016; Gentes et al. 2017; Lazaro et al. 2018), but recent work emphasized the importance of considering diel periphytic dynamics in MeHg production (Bouchet et al. 2018).
Currently, there is very little support for phototrophs participating directly in Hg methylation. Aside from the initial research cited in our first review (Pongratz and Heumann 1998; Deng et al. 2013), only one new study has emerged investigating the potential for direct methylation in the cyanobacterium Nostoc paludosum that showed no MeHg production (Franco et al. 2018).
Microbial strains in the phylum Chloroflexi, which includes representatives of the anoxygenic phototrophic green non-sulphur bacteria, are now known to host the hgcAB gene cluster. However, the gene sequences found to date belong to the chemotrophic Dehalococcoidaceae family (Bae et al. 2014; Schaefer et al. 2014a; Podar et al. 2015). With the increasing diversity of habitats being investigated for Hg methylation, perhaps a phototrophic representative harbouring the hgcAB gene cluster capable of methylation may soon be uncovered.
Phototrophic methylmercury demethylation
Previously, there was little to no mechanistic data available for direct phototroph-mediated MeHg demethylation (Grégoire and Poulain 2014). Recently, Kritee et al. 2017 used Hg stable isotope fractionation in Isochrysis galbana to demonstrate MeHg demethylation in algal cells. In this study, cells preferentially demethylated lighter MeHg isotopes resulting in a pool of isotopically light HgII (Kritee et al. 2017). In addition, MeHg demethylation resulted in a positive odd-isotope mass-independent signature that was characteristic of the photochemical destruction of MeHg and similar to the Hg isotope signature observed in a variety of marine and freshwater fish (Kritee et al. 2017). This study highlights the power of Hg stable isotope fractionation to track phototroph-mediated Hg transformations at the environmental scale.
Phototrophic mercury reduction
Most of the previous evidence for phototroph-mediated Hg reduction stemmed from observations and experiments performed with oxygenic phototrophs (Grégoire and Poulain 2014). Recent work on anoxygenic phototrophic PNSB (Grégoire and Poulain 2016) and Heliobacteria (Grégoire et al. 2018) has provided some of the first mechanistic details for anaerobic phototrophic Hg reduction.
Our work with PNSB demonstrated that HgII was preferentially reduced during photoheterotrophic growth on reduced organic carbon substrate (e.g., using light as an energy source and generating biomass using an organic carbon source) (Grégoire and Poulain 2016). Under controlled laboratory conditions, we showed that PNSB derived an advantage from sublethal Hg exposure, which we attributed to the ability of Hg to act as an electron sink during phototrophic growth (Grégoire and Poulain 2016). Large peaks of Hg0 have been reported in the metalimnion of lakes (Poulain et al. 2004), and anoxygenic phototrophs such as PNSB thrive in environments such as the thermoclines and chemoclines of stratified aquatic ecosystems (Schütte et al. 2016) and microbial mats (Schneider et al. 2013). These metabolically versatile microbes stand to be important players in Hg redox cycling in environments where light is attenuated and reduced organic carbon is available. Such habitats are often located at redox interfaces that act as gateways to Hg methylation sites.
Following up on our work with anoxygenic phototrophs, we further discovered that Heliobacteria, a family of spore-forming fermentative photoheterotrophs of the order Clostridiales, were among the most efficient Hg reducers reported to date (Grégoire et al. 2018). No apparent dedicated Hg reduction machinery (i.e., mer operon determinants) was found in the genome of Heliobacterium modesticaldum Ice1, and HgII reduction most likely occurred as a cometabolic process related to the availability of reduced redox cofactors (e.g., ferredoxin) (Grégoire et al. 2018). The fact that cometabolic Hg reduction was observed in two phylogenetically and ecologically distinct groups of anoxygenic phototrophs suggest that such pathways may be widespread in anoxic habitats offering phototrophs a means of detoxifying Hg without dedicated enzymatic machinery.
The use of stable Hg isotopes has recently provided unique insights into oxygenic phototrophic Hg reduction. In the same study addressing MeHg demethylation in I. galbana, cells reduced lighter HgII isotopes with the strongest fractionation signature occurring within the intracellular pool of HgII (Kritee et al. 2017). The authors also observed mass-independent fractionation signatures suggesting that Hg was reduced by free radicals inside the cell following binding to thiol functional groups (Zheng and Hintelmann 2010; Kritee et al. 2017). The prevalence of free radical formation during oxygenic photosynthesis is such that algal cells could be seen as small photoreactors hosting Hg photoreduction. Indeed, this study represents a fascinating case of intracellular metal photoreduction that blurs the lines between abiotic and biotic transformations.
The presence of putative merA homologues in the genomes of some anoxygenic phototrophs (Mukkata et al. 2015; Pérez et al. 2018) highlights an interesting question regarding Hg tolerance in phototrophs: do cells preferentially detoxify Hg via cometabolic processes or through the use of dedicated reductases? Currently, it seems that phototrophic Hg reduction occurs mostly as a cometabolic process that relies on core metabolic machinery rather than mer-like determinants, which are largely absent in phototrophs. That being said, very few studies have been published on the subject and the presence of such pathways has only been identified for a small number of model organisms. By exploring this question further we will better define the physiological mechanisms supporting phototrophic Hg metabolism. The increasing availability of microbial genomes and genetic tractability of anoxygenic phototrophs should help address this question in future research.
Clearly, phototrophs are more than just an entry point for Hg into food webs. The mechanistic findings mentioned in this section frame photosynthetic redox homeostasis as a metabolic process coupled with Hg reduction. These mechanistic studies further demonstrate that phototrophs can directly impact the substrate of Hg available to methylators via Hg redox cycling. How such interactions play out in the environment remains elusive, however.
Phototrophic mercury oxidation
At present, there is a lack of studies addressing Hg0 oxidation in phototrophs. This could be related to the challenges associated with working with volatile Hg0 as a substrate. We speculate that anoxygenic phototrophic bacteria could potentially oxidize Hg0 through thiol-related mechanisms similar to those outlined in chemotrophic anaerobes based on the fact that Hg is known to bind to thiols in the photosynthetic reaction center of PNSB (Asztalos et al. 2012; Sipka et al. 2017). Currently, the presence of such pathways has yet to be investigated and Hg0 oxidation in phototrophs has yet to be demonstrated.
Mercury toxicity to phototrophs
Phototrophs appear to mitigate Hg toxicity either via intracellular sequestration or elimination from the cell (e.g., via efflux or evasion). Sequestration strategies come in the form of dedicated chelating molecules such as phytochelatins and metallothioneins (Rauser 1990; Kawakami et al. 2006) and the production of HgS (Kelly et al. 2007), all of which prevent Hg from binding to sensitive target sites inside the cell.
Inside the cell, Hg can interact with a number of proteins that are essential for supporting photosynthetic metabolism. Hg’s toxicity is attributable to Hg’s affinity for thiol bonds (Rooney 2007) and its ability to substitute for cations that serve as cofactors in enzymatic reactions essential to capturing light energy (Matson et al. 1972; Singh et al. 2012; Zhang et al. 2013). By disrupting protein function, Hg can inhibit electron transport and the ability of cells to generate ATP thereby inhibiting cell growth (Murthy and Mohanty 1993; Kukarskikh et al. 2003; Antal et al. 2009). Although toxicological studies continue to emerge evaluating the ranges of Hg concentrations tolerated by different model phototrophs (Chen et al. 2014; Zhu et al. 2015; Nowicka et al. 2016; Mu et al. 2017), very few offer new mechanistic insights (Grégoire and Poulain 2014).
The recent transcriptomic work with C. reinhardtii has provided insights into the genetic response of green algae to Hg toxicity (Beauvais-Fluck et al. 2016; Beauvais-Fluck et al. 2017). Hg exposure led to the disruption of genes involved in motility, cell division, energy metabolism, amino acid production, lipid oxidation, metal transport, and antioxidant enzyme synthesis (Beauvais-Fluck et al. 2017). Although the apparent negative effects on gene regulation did not always manifest as a physiological response (Beauvais-Fluck et al. 2016; Beauvais-Fluck et al. 2017), the authors saw increased photosynthetic efficiency when small levels of MeHg were present (Beauvais-Fluck et al. 2016). These results suggested a possible hormetic effect derived from MeHg exposure that merits further investigation. Based on our own observations in PNSB, there is precedence for sublethal exposure to Hg providing a physiological advantage during phototrophic growth by acting as an electron sink (Grégoire and Poulain 2016). Whether such a pathway is supported in oxygenic phototrophs has yet to be tested and the importance of beneficial or hormetic responses to Hg in the environment remains to be evaluated.
At the time of publishing our 2014 review paper, very little mechanistic information was available on Hg toxicity in anoxygenic phototrophs. This has changed in recent years. Work with PNSB showed that large amounts of HgII can bind to the photosynthetic reaction center (PS-RC) (Asztalos et al. 2012) and that the vast majority of Hg supplied to PNSB was bound to weak binding sites, rather than to high affinity thiol-bearing sites (Sipka et al. 2017). Although the exact chemical natures of the weak binding sites were not further discussed, one can speculate that these binding sites may have been carboxyl groups that have a lower affinity for Hg (Gu et al. 2011; Mishra et al. 2011; Jiang et al. 2015) and are known to be present in the PS-RC (Knox et al. 2018). Sipka et al. 2017 also demonstrated that Hg toxicity was attributable to a small number of strong thiol-bearing Hg binding sites that disrupted cyclical phosphorylation by inhibiting inter-quinone electron transfer and proton translocation. Despite the sensitivity of electron transport machinery to Hg, model PNSB strains could tolerate μM levels of Hg before damage to photosynthetic membranes became apparent (Kis et al. 2015).
These new findings illustrate that Hg targets may be similar between oxygenic anoxygenic phototrophs, which is unsurprising given the evolutionary parallels between some components of their photosynthetic machinery. Although the availability of genomic information frame PNSB as excellent models to study Hg toxicity in anoxygenic phototrophs, the response of other clades to Hg has yet to be characterized. For instance, we know very little about Hg interactions with green (non)-sulphur bacteria, aerobic anoxygenic photosynthetic bacteria, Heliobacteria, or purple sulphur bacteria. This information would be useful in revealing the evolutionary and physiological context shaping the response to Hg toxicity among phylogenetically distinct clades of phototrophs. Indeed, the absence of dedicated Hg resistance strategies such as the mer operon among anaerobes and phototrophs is puzzling. There appears to be a tight coupling between the presence of oxygen and the evolution of the mer operon (Barkay et al. 2010). Anaerobic and anoxygenic phototrophic metabolisms predate the rise of oxygen on Earth and cometabolic processes such as sequestration or the use of excess reducing power to catalyze Hg reduction may have been sufficient to cope with Hg toxicity in the absence of oxygen.
Concluding remarks
Over the last four years, we have observed a shift in the field of research addressing microbial Hg transformations. Investigations into chemotrophic Hg methylation still dominate the literature thanks largely to the discovery of the hgcAB gene cluster. With powerful molecular tools in hand, one important frontier in our field is to identify the native function of this gene cluster and the possible role(s) associated with Hg methylation. In addition to gaining fundamental insights into the process, such mechanistic insights may reveal strategies that could help manage Hg pollution.
Important mechanistic details of anaerobic and phototrophic Hg cycling have also emerged, leading us to reconsider the environmental contributions of anaerobic Hg redox cycling pathways. One such contribution is summarized in Fig. 1, highlighting a possible role of phototrophs in catalyzing Hg reduction at redox interfaces within a stratified lake ecosystem.
Fig. 1.
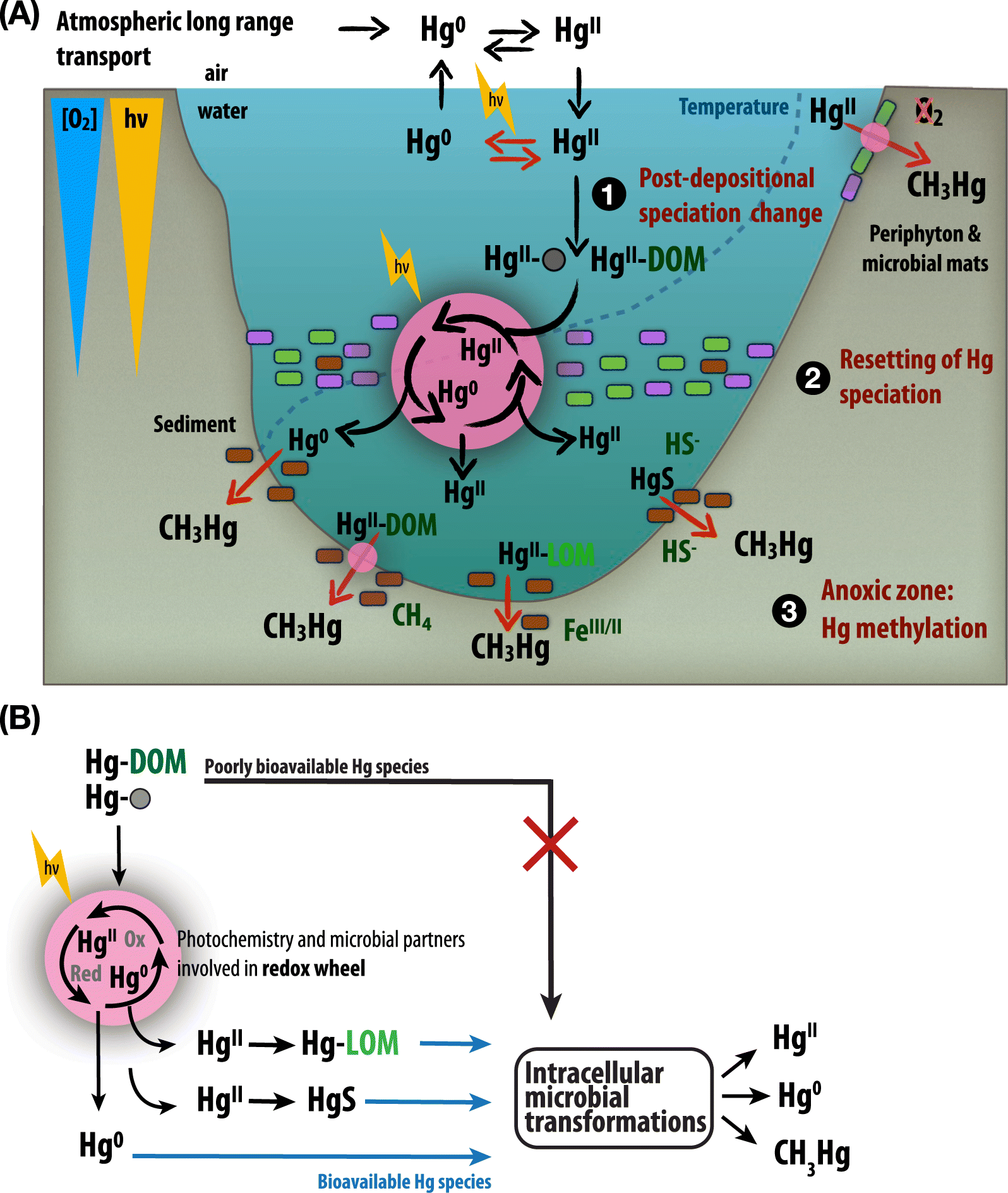
Once deposited into aquatic ecosystems, aqueous Hg speciation is altered such that ageing of Hg complexes occurs, typically leading to low Hg bioavailability over time (Fig. 1A-(1)). Redox processes catalyzing the transformation of Hg complexes control the delivery of bioavailable Hg to anoxic zones. This occurs by resetting Hg speciation, i.e., remobilizing Hg initially present as poorly bioavailable complexes (e.g., Hg-DOM or Hg-particles) to more mobile and bioavailable species (e.g., Hg0, Fig. 1A-(2) and 1B). This can occur by anaerobic or photobiological redox processes targeting Hg and described in this review, or via heterotrophic or photochemical degradation of the organic ligands to which Hg is bound. Given the increasing importance of anaerobes in Hg0 oxidation and anoxygenic phototrophs in HgII reduction, these groups can provide a fresh supply of bioavailable Hg to sites conducive to Hg methylation (Fig. 1A-(3) and 1B).
This conceptual redox wheel (Chiasson-Gould et al. 2014) is meant to highlight the potential for highly dynamic yet cryptic Hg redox cycling (Fig. 1B). We use the word cryptic here to highlight the fact that no net Hg0 accumulation may be observed but its production and rapid oxidation may represent an essential step in controlling Hg mobility at, and across, redox interfaces. Furthermore, this conceptual framework illustrates how phototrophs (or other anaerobes) can participate in Hg transformation well beyond their role as MeHg accumulators at the base of food webs. Note that such processes can occur in any stratified environment, whether it is within the metalimnion of lakes, microbial mats, or periphytic communities (Fig. 1).
Although lab studies have afforded the controlled conditions necessary to elucidate new mechanisms for phototrophic Hg transformations, these pathways continue to be understudied in the environment. This knowledge gap will become increasingly important to address in the face of predicted environmental change. Increases in temperature, prolonged ice-free seasons and more intense water column stratification will have a dramatic impact on phototroph dynamics that, in turn, exert important controls on environmental chemistry and metal speciation. Indeed, there stand to be far-reaching impacts on global Hg cycling stemming from changes in phototrophic communities that occupy broad niches in oxic and anoxic habitats.
Acknowledgements
This paper has been improved thanks to the work of three reviewers to whom we are grateful. We also would like to thank Tamar Barkay and Britt D. Hall for comments on an earlier version of this manuscript and Cindy Gilmour for her continued insights on the role of microbes in Hg cycling. Our work was funded by NSERC Discovery and Accelerator grants, CFI funding to AJP, and NSERC graduate scholarship to DSG.
References
Antal TK, Graevskaya EE, Matorin DN, Volgusheva AA, Osipov VA, Krendeleva TE, et al. 2009. Assessment of the effects of methylmercury and copper ions on primary processes of photosynthesis in green microalga Chlamydomonas moewusii by analysis of the kinetic curves of variable chlorophyll fluorescence. Biophysics, 54: 481–485.
Asztalos E, Sipka G, Kis M, Trotta M, and Maróti P. 2012. The reaction center is the sensitive target of the mercury(II) ion in intact cells of photosynthetic bacteria. Photosynthesis Research, 112: 129–140.
Bae HS, Dierberg FE, and Ogram A. 2014. Syntrophs dominate sequences associated with the mercury methylation-related gene hgcA in the water conservation areas of the Florida Everglades. Applied and Environmental Microbiology, 80: 6517–6526.
Baral BS, Bandow NL, Vorobev A, Freemeier BC, Bergman BH, Herdendorf TJ, et al. 2014. Mercury binding by methanobactin from Methylocystis strain SB2. Journal of Inorganic Biochemistry, 141: 161–169.
Barkay T, and Wagner-Dobler I. 2005. Microbial transformations of mercury: potentials, challenges, and achievements in controlling mercury toxicity in the environment. In Advances in applied microbiology. Edited by AI Laskin, JW Bennett, and GM Gadd. Elsevier Academic Press Inc., San Diego, California. pp. 1–52.
Barkay T, Miller SM, and Summers AO. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiology Reviews, 27: 355–384.
Barkay T, Kritee K, Boyd E, and Geesey G. 2010. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environmental Microbiology, 12: 2904–2917.
Beauvais-Flück R, Slaveykova VI, and Cosio C. 2016. Transcriptomic and physiological responses of the green microalga Chlamydomonas reinhardtii during short-term exposure to subnanomolar methylmercury concentrations. Environmental Science & Technology, 50: 7126–7134.
Beauvais-Flück R, Slaveykova VI, and Cosio C. 2017. Cellular toxicity pathways of inorganic and methyl mercury in the green microalga Chlamydomonas reinhardtii. Scientific Reports, 7:
Bouchet S, Goñi-Urriza M, Monperrus M, Guyoneaud R, Fernandez P, Heredia C, et al. 2018. Linking microbial activities and low molecular weight thiols to Hg methylation in biofilms and periphyton from high altitude tropical lakes in the Bolivian altiplano. Environmental Science & Technology.
Bouffard A, and Amyot M. 2009. Importance of elemental mercury in lake sediments. Chemosphere, 74: 1098–1103.
Bravo AG, Loizeau JL, Dranguet P, Makri S, Björn E, Ungureanu VG, et al. 2016. Persistent Hg contamination and occurrence of Hg-methylating transcript (hgcA) downstream of a chlor-alkali plant in the Olt River (Romania). Environmental Science and Pollution Research, 23: 10529–10541.
Bravo AG, Zopfi J, Buck M, Xu J, Bertilsson S, Schaefer JK, et al. 2018. Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by waste water releases. The ISME Journal, 12: 802–812.
Cárdenas CS, Diéguez MC, Guevara SR, Marvin-Dipasquale M, and Queimaliños CP. 2014. Incorporation of inorganic mercury (Hg2+) in pelagic food webs of ultraoligotrophic and oligotrophic lakes: the role of different plankton size fractions and species assemblages. Science of the Total Environment, 494: 65–73.
Chen H-W, Huang W-J, Wu T-H, and Hon C-L. 2014. Effects of extracellular polymeric substances on the bioaccumulation of mercury and its toxicity toward the cyanobacterium Microcystis aeruginosa. Journal of Environmental Science and Health – Part A Toxic/Hazardous Substances and Environmental Engineering, 49: 1370–1379.
Chiasson-Gould SA, Blais JM, and Poulain AJ. 2014. Dissolved organic matter kinetically controls mercury bioavailability to bacteria. Environmental Science & Technology, 48: 3153–3161.
Choi SC, Chase T Jr, and Bartha R. 1994a. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Applied and Environmental Microbiology, 60: 1342–1346.
Choi SC, Chase T Jr, and Bartha R. 1994b. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Applied and Environmental Microbiology, 60: 4072–4077.
Christensen GA, Wymore AM, King AJ, Podar M, Hurt RA, Santillan EU, et al. 2016. Development and validation of broad-range qualitative and clade-specific quantitative molecular probes for assessing mercury methylation in the environment. Applied and Environmental Microbiology, 82: 6068–6078.
Christensen GA, Somenahally AC, Moberly JG, Miller CM, King AJ, Gilmour CC, et al. 2018. Carbon amendments alter microbial community structure and net mercury methylation potential in sediments. Applied and Environmental Microbiology, 84: e01049.
Colombo MJ, Ha J, Reinfelder JR, Barkay T, and Yee N. 2013. Anaerobic oxidation of Hg(0) and methylmercury formation by Desulfovibrio desulfuricans ND132. Geochimica et Cosmochimica Acta, 112: 166–177.
Colombo MJ, Ha J, Reinfelder JR, Barkay T, and Yee N. 2014. Oxidation of Hg(0) to Hg(II) by diverse anaerobic bacteria. Chemical Geology, 363: 334–340.
Deng GF, Zhang TW, Yang LM, and Wang QQ. 2013. Studies of biouptake and transformation of mercury by a typical unicellular diatom Phaeodactylum tricornutum. Chinese Science Bulletin, 58: 256–265.
Desrochers KAN, Paulson KMA, Ptacek CJ, Blowes DW, and Gould WD. 2015. Effect of electron donor to sulfate ratio on mercury methylation in floodplain sediments under saturated flow conditions. Geomicrobiology Journal, 32: 924–933.
Dranguet P, Le Faucheur S, Cosio C, and Slaveykova VI. 2017. Influence of chemical speciation and biofilm composition on mercury accumulation by freshwater biofilms. Environmental Science-Processes & Impacts, 19: 38–49.
Dupraz C, and Visscher PT. 2005. Microbial lithification in marine stromatolites and hypersaline mats. Trends in Microbiology, 13: 429–438.
Fleming EJ, Mack EE, Green PG, and Nelson DC. 2006. Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Applied and Environmental Microbiology, 72: 457–464.
Franco MW, Mendes LA, Windmöller CC, Moura KAF, Oliveira LAG, and Barbosa FAR. 2018. Mercury methylation capacity and removal of Hg species from aqueous medium by cyanobacteria. Water, Air, & Soil Pollution, 229: 127.
Gentes S, Taupiac J, Colin Y, Andre JM, and Guyoneaud R. 2017. Bacterial periphytic communities related to mercury methylation within aquatic plant roots from a temperate freshwater lake (South-Western France). Environmental Science and Pollution Research, 24: 19223–19233.
Gilmour CC, Bullock AL, McBurney A, Podar M, and Elias DA. 2018. Robust mercury methylation across diverse methanogenic Archaea. mBio, 9: e02403.
Gionfriddo CM, Tate MT, Wick RR, Schultz MB, Zemla A, Thelen MP, et al. 2016. Microbial mercury methylation in Antarctic sea ice. Nature Microbiology, 1: 16127.
Graham AM, Aiken GR, and Gilmour CC. 2013. Effect of dissolved organic matter source and character on microbial Hg methylation in Hg-S-DOM solutions. Environmental Science & Technology, 47: 5746–5754.
Graham AM, Cameron-Burr KT, Hajic HA, Lee C, Msekela D, and Gilmour CC. 2017. Sulfurization of dissolved organic matter increases Hg-sulfide-dissolved organic matter bioavailability to a Hg-methylating bacterium. Environmental Science & Technology, 51: 9080–9088.
Graham EB, Gabor RS, Schooler S, Mcknight DM, Nemergut DR, and Knelman JE. 2018. Oligotrophic wetland sediments susceptible to shifts in microbiomes and mercury cycling with dissolved organic matter addition. PeerJ, 6: e4575.
Grégoire DS, and Poulain AJ. 2014. A little bit of light goes a long way: the role of phototrophs on mercury cycling. Metallomics, 6: 396–407.
Grégoire DS, and Poulain AJ. 2016. A physiological role for Hg(II) during phototrophic growth. Nature Geoscience, 9: 121–125.
Grégoire DS, Lavoie NC, and Poulain AJ. 2018. Heliobacteria reveal fermentation as a key pathway for mercury reduction in anoxic environments. Environmental Science & Technology, 52: 4145–4153.
Gu B, Bian Y, Miller CL, Dong W, Jiang X, and Liang L. 2011. Mercury reduction and complexation by natural organic matter in anoxic environments. Proceedings of the National Academy of Sciences of the USA, 108: 1479–1483.
Hamelin S, Amyot M, Barkay T, Wang Y, and Planas D. 2011. Methanogens: principal methylators of mercury in lake periphyton. Environmental Science & Technology, 45: 7693–7700.
Han S, Narasingarao P, Obraztsova A, Gieskes J, Hartmann AC, Tebo BM, et al. 2010. Mercury speciation in marine sediments under sulfate-limited conditions. Environmental Science & Technology, 44: 3752–3757.
Hsu-Kim H, Kucharzyk KH, Zhang T, and Deshusses MA. 2013. Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: a critical review. Environmental Science & Technology, 47: 2441–2456.
Janssen SE, Schaefer JK, Barkay T, and Reinfelder JR. 2016. Fractionation of mercury stable isotopes during microbial methylmercury production by iron- and sulfate-reducing bacteria. Environmental Science & Technology, 50: 8077–8083.
Jiang T, Skyllberg U, Wei SQ, Wang DY, Lu S, Jiang ZM, et al. 2015. Modeling of the structure-specific kinetics of abiotic, dark reduction of Hg(II) complexed by O/N and S functional groups in humic acids while accounting for time-dependent structural rearrangement. Geochimica et Cosmochimica Acta, 154: 151–167.
Kawakami SK, Gledhill M, and Achterberg EP. 2006. Production of phytochelatins and glutathione by marine phytoplankton in response to metal stress. Journal of Phycology, 42: 975–989.
Kelly DJA, Budd K, and Lefebvre DD. 2007. Biotransformation of mercury in pH-stat cultures of eukaryotic freshwater algae. Archives of Microbiology, 187: 45–53.
Kis M, Sipka G, and Maróti P. 2017. Stoichiometry and kinetics of mercury uptake by photosynthetic bacteria. Photosynthesis Research, 132: 197–209.
Kis M, Sipka G, Asztalos E, Rázga Z, and Maróti P. 2015. Purple non-sulfur photosynthetic bacteria monitor environmental stresses. Journal of Photochemistry and Photobiology B: Biology, 151: 110–117.
Knox PP, Gorokhov VV, Korvatovskiy BN, Lukashev EP, Goryachev SN, Paschenko VZ, et al. 2018. The effect of light and temperature on the dynamic state of Rhodobacter sphaeroides reaction centers proteins determined from changes in tryptophan fluorescence lifetime and P+QA− recombination kinetics. Journal of Photochemistry and Photobiology B-Biology, 180: 140–148.
Krabbenhoft DP, and Sunderland EM. 2013. Global change and mercury. Science, 341: 1457–1458.
Kritee K, Motta LC, Blum JD, Tsui MT-K, and Reinfelder JR. 2017. Photomicrobial visible light-induced magnetic mass independent fractionation of mercury in a marine microalga. ACS Earth and Space Chemistry, 2(5): 432–440.
Kronberg R-M, Schaefer JK, Björn E, and Skyllberg U. 2018. Mechanisms of methylmercury net degradation in alder swamps: the role of methanogens and abiotic processes. Environmental Science & Technology Letters, 5(4): 220–225.
Kukarskikh GP, Graevskaya EE, Krendeleva TE, Timofeev KN, and Rubin AB. 2003. Effect of methylmercury on primary photosynthesis processes in green microalgae Chlamydomonas reinhardtii. Biophysics, 48(5): 853–859.
Landner L. 1971. Biochemical model for the biological methylation of mercury suggested from methylation studies in vivo with Neurospora crassa. Nature, 230: 452–454.
Lanza WG, Achá D, Point D, Masbou J, Alanoca L, Amouroux D, and Lazzaro X. 2017. Association of a specific algal group with methylmercury accumulation in periphyton of a tropical high-altitude Andean lake. Archives of Environmental Contamination and Toxicology, 72: 1–10.
Lázaro WL, Díez S, Da Silva CJ, Ignácio ARA, and Guimarães JRD. 2018. Seasonal changes in peryphytic microbial metabolism determining mercury methylation in a tropical wetland. Science of the Total Environment, 627: 1345–1352.
Lee S, Kim DH, and Kim KW. 2018. The enhancement and inhibition of mercury reduction by natural organic matter in the presence of Shewanella oneidensis MR-1. Chemosphere, 194: 515–522.
Lin H, Hurt RA, Johs A, Parks JM, Morrell-Falvey JL, Liang L, et al. 2014a. Unexpected effects of gene deletion on interactions of mercury with the methylation-deficient mutant ΔhgcAB. Environmental Science and Technology Letters, 1: 271–276.
Lin H, Morrell-Falvey JL, Rao B, Liang L, and Gu B. 2014b. Coupled mercury-cell sorption, reduction, and oxidation on methylmercury production by Geobacter sulfurreducens PCA. Environmental Science & Technology, 48: 11969–11976.
Lin H, Lu X, Liang LY, and Gu BH. 2015. Cysteine inhibits mercury methylation by Geobacter sulfurreducens PCA mutant ΔomcBESTZ. Environmental Science & Technology Letters, 2: 144–148.
Liu J, Valsaraj KT, and Delaune RD. 2009. Inhibition of mercury methylation by iron sulfides in an anoxic sediment. Environmental Engineering Science, 26: 833–840.
Liu S, and Wiatrowski HA. 2018. Reduction of Hg(II) to Hg(0) by biogenic magnetite from two magnetotactic bacteria. Geomicrobiology Journal, 35: 198–208.
Liu YR, Lu X, Zhao LD, An J, He JZ, Pierce EM, et al. 2016. Effects of cellular sorption on mercury bioavailability and methylmercury production by Desulfovibrio desulfuricans ND132. Environmental Science & Technology, 50: 13335–13341.
Lu X, Liu Y, Johs A, Zhao L, Wang T, Yang Z, et al. 2016. Anaerobic mercury methylation and demethylation by Geobacter bemidjiensis Bem. Environmental Science & Technology, 50: 4366–4373.
Lu X, Gu WY, Zhao LD, Ul Haque MF, Dispirito AA, Semrau JD, et al. 2017. Methylmercury uptake and degradation by methanotrophs. Science Advances, 3(5): e1700041.
Ma M, Du H, Wang D, and Sun T. 2017a. Mercury methylation in the soils and sediments of Three Gorges Reservoir Region. Journal of Soils and Sediments, 18(3): 1100–1109.
Ma M, Du HX, Wang DY, Kang SC, and Sun T. 2017b. Biotically mediated mercury methylation in the soils and sediments of Nam Co Lake, Tibetan Plateau. Environmental Pollution, 227: 243–251.
Mahbub KR, Krishnan K, Naidu R, Andrews S, and Megharaj M. 2017. Mercury toxicity to terrestrial biota. Ecological Indicators, 74: 451–462.
Matson RS, Mustoe GE, and Chang SB. 1972. Mercury inhibition on lipid biosynthesis in freshwater algae. Environmental Science & Technology, 6: 158–160.
Mazrui NM, Seelen E, King’ondu CK, Thota S, Awino J, Rouge J, et al. 2018. The precipitation, growth and stability of mercury sulfide nanoparticles formed in the presence of marine dissolved organic matter. Environmental Science: Processes & Impacts, 20: 642–656.
Mazur MEE, Eckley CS, and Mitchell CPJ. 2015. Susceptibility of soil bound mercury to gaseous emission as a function of source depth: an enriched isotope tracer investigation. Environmental Science & Technology, 49: 9143–9149.
Mishra B, O’Loughlin EJ, Boyanov MI, and Kemner KM. 2011. Binding of HgII to high-affinity sites on bacteria inhibits reduction to Hg0 by mixed FeII/III phases. Environmental Science & Technology, 45: 9597–9603.
Mu W, Jia K, Liu Y, Pan X, and Fan Y. 2017. Response of the freshwater diatom Halamphora veneta (Kützing) Levkov to copper and mercury and its potential for bioassessment of heavy metal toxicity in aquatic habitats. Environmental Science and Pollution Research, 24: 26375–26386.
Mukkata K, Kantachote D, Wittayaweerasak B, Techkarnjanaruk S, Mallavarapu M, and Naidu R. 2015. Distribution of mercury in shrimp ponds and volatilization of Hg by isolated resistant purple nonsulfur bacteria. Water, Air, and Soil Pollution, 226: 148.
Murthy SDS, and Mohanty P. 1993. Mercury ions inhibit photosynthetic electron transport at multiple sites in the cyanobacterium Synechococcus 6301. Journal of Biosciences, 18: 355–360.
Ndu U, Barkay T, Schartup AT, Mason RP, and Reinfelder JR. 2016. The effect of aqueous speciation and cellular ligand binding on the biotransformation and bioavailability of methylmercury in mercury-resistant bacteria. Biodegradation, 27: 29–36.
Norambuena J, Wang Y, Hanson T, Boyd JM, and Barkay T. 2018. Low-molecular-weight thiols and thioredoxins are important players in Hg(II) resistance in Thermus thermophilus HB27. Applied and Environmental Microbiology, 84.
Nowicka B, Pluciński B, Kuczyńska P, and Kruk J. 2016. Physiological characterization of Chlamydomonas reinhardtii acclimated to chronic stress induced by Ag, Cd, Cr, Cu and Hg ions. Ecotoxicology and Environmental Safety, 130: 133–145.
Obrist D, Kirk JL, Zhang L, Sunderland EM, Jiskra M, and Selin NE. 2018. A review of global environmental mercury processes in response to human and natural perturbations: changes of emissions, climate, and land use. Ambio, 47: 116–140.
Olsen TA, Brandt CC, and Brooks SC. 2016. Periphyton biofilms influence net methylmercury production in an industrially contaminated system. Environmental Science & Technology, 50: 10843–10850.
Oremland RS, Culbertson CW, and Winfrey MR. 1991. Methylmercury decomposition in sediments and bacterial cultures: involvement of methanogens and sulfate reducers in oxidative demethylation. Applied and Environmental Microbiology, 57: 130–137.
Paranjape AR, and Hall BD. 2017. Recent advances in the study of mercury methylation in aquatic systems. FACETS, 2: 85–119.
Parks JM, and Smith JC. 2016. Modeling mercury in proteins. In Computational approaches for studying enzyme mechanism, Part B. Edited by GA Voth. Academic Press, Elsevier Inc., San Diego, California. pp. 103–122.
Parks JM, Guo H, Momany C, Liang L, Miller SM, Summers AO, et al. 2009. Mechanism of Hg–C protonolysis in the organomercurial lyase MerB. Journal of the American Chemical Society, 131: 13278–13285.
Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr, Smith SD, et al. 2013. The genetic basis for bacterial mercury methylation. Science, 339: 1332–1335.
Pérez V, Dorador C, Molina V, Yáñez C, and Hengst M. 2018. Rhodobacter sp. Rb3, an aerobic anoxygenic phototroph which thrives in the polyextreme ecosystem of the Salar de Huasco, in the Chilean Altiplano. Antonie van Leeuwenhoek, 111(8): 1449–1465.
Pham AL-T, Morris A, Zhang T, Ticknor J, Levard C, and Hsu-Kim H. 2014. Precipitation of nanoscale mercuric sulfides in the presence of natural organic matter: structural properties, aggregation, and biotransformation. Geochimica et Cosmochimica Acta, 133: 204–215.
Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR, et al. 2015. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Science Advances, 1: e1500675–e1500675.
Polerecky L, Bachar A, Schoon R, Grinstein M, Jorgensen BB, De Beer D, et al. 2007. Contribution of Chloroflexus respiration to oxygen cycling in a hypersaline microbial mat from Lake Chiprana, Spain. Environmental Microbiology, 9: 2007–2024.
Pongratz R, and Heumann KG. 1998. Production of methylated mercury and lead by polar macroalgae – a significant natural source for atmospheric heavy metals in clean room compartments. Chemosphere, 36: 1935–1946.
Poulain AJ, Amyot M, Findlay D, Telor S, Barkay T, and Hintelmann H. 2004. Biological and photochemical production of dissolved gaseous mercury in a boreal lake. Limnology and Oceanography, 49: 2265–2275.
Poulin BA, Aiken GR, Nagy KL, Monceau A, Krabbenhoft DP, and Ryan JN. 2016. Mercury transformation and release differs with depth and time in a contaminated riparian soil during simulated flooding. Geochimica Et Cosmochimica Acta, 176: 118–138.
Qian C, Johs A, Chen H, Mann BF, Lu X, Abraham PE, et al. 2016. Global proteome response to deletion of genes related to mercury methylation and dissimilatory metal reduction reveals changes in respiratory metabolism in Geobacter sulfurreducens PCA. Journal of Proteome Research, 15: 3540–3549.
Rauser WE. 1990. Phytochelatins. Annual Review of Biochemistry, 59: 61–86.
Raven JA. 2009. Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquatic Microbial Ecology, 56: 177–192.
Rooney JPK. 2007. The role of thiols, dithiols, nutritional factors and interacting ligands in the toxicology of mercury. Toxicology, 234: 145–156.
Rossi F, Potrafka RM, Pichel FG, and De Philippis R. 2012. The role of the exopolysaccharides in enhancing hydraulic conductivity of biological soil crusts. Soil Biology & Biochemistry, 46: 33–40.
Schaefer JK, Kronberg R-M, Morel FMM, and Skyllberg U. 2014a. Detection of a key Hg methylation gene, hgcA, in wetland soils. Environmental Microbiology Reports, 6: 441–447.
Schaefer JK, Szczuka A, and Morel FMM. 2014b. Effect of divalent metals on Hg(II) uptake and methylation by bacteria. Environmental Science & Technology, 48: 3007–3013.
Schneider D, Arp G, Reimer A, Reitner J, and Daniel R. 2013. Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati Atoll, Central Pacific. PLoS ONE, 8(6): e66662.
Schütte UME, Cadieux SB, Hemmerich C, Pratt LM, and White JR. 2016. Unanticipated geochemical and microbial community structure under seasonal ice cover in a dilute, dimictic Arctic lake. Frontiers in Microbiology, 7: 1035.
Selin NE, Jacob DJ, Park RJ, Yantosca RM, Strode S, Jaegle L, et al. 2007. Chemical cycling and deposition of atmospheric mercury: global constraints from observations. Journal of Geophysical Research: Atmospheres, 112: D02308.
Silva PJ, and Rodrigues V. 2015. Mechanistic pathways of mercury removal from the organomercurial lyase active site. PeerJ, 3: e1127.
Singh R, Srivastava PK, Singh VP, Dubey G, and Prasad SM. 2012. Light intensity determines the extent of mercury toxicity in the cyanobacterium Nostoc muscorum. Acta Physiologiae Plantarum, 34: 1119–1131.
Sipka G, Kis M, and Maróti P. 2017. Characterization of mercury(II)-induced inhibition of photochemistry in the reaction center of photosynthetic bacteria. Photosynthesis Research, 136(3): 379–392.
Sloth NP, Riemann B, Nielsen LP, and Blackburn TH. 1996. Resilience of pelagic and benthic microbial communities to sediment resuspension in a coastal ecosystem, Knebel Vig, Denmark. Estuarine Coastal and Shelf Science, 42: 405–415.
Soto Cárdenas C, Gerea M, Queimaliños C, Ribeiro Guevara S, and Diéguez MC. 2018. Inorganic mercury (Hg2+) accumulation in autotrophic and mixotrophic planktonic protists: implications for Hg trophodynamics in ultraoligotrophic Andean Patagonian lakes. Chemosphere, 199: 223–231.
Stenzler B, Hinz A, Ruuskanen M, and Poulain AJ. 2017. Ionic strength differentially affects the bioavailability of neutral and negatively charged inorganic Hg complexes. Environmental Science & Technology, 51: 9653–9662.
Su Y-B, Chang W-C, Hsi H-C, and Lin C-C. 2016. Investigation of biogeochemical controls on the formation, uptake and accumulation of methylmercury in rice paddies in the vicinity of a coal-fired power plant and a municipal solid waste incinerator in Taiwan. Chemosphere, 154: 375–384.
Sunderland EM. 2007. Mercury exposure from domestic and imported estuarine and marine fish in the U.S. seafood market. Environmental Health Perspectives, 115: 235–242.
Vazquez-Rodriguez AI, Hansel CM, Zhang T, Lamborg CH, Santelli CM, Webb SM, et al. 2015. Microbial- and thiosulfate-mediated dissolution of mercury sulfide minerals and transformation to gaseous mercury. Frontiers in Microbiology, 6: 596.
Vishnivetskaya TA, Hu H, Van Nostrand JD, Wymore A, Xu X, Qiu G, et al. 2018. Microbial community structure with trends in methylation gene diversity and abundance in mercury-contaminated rice paddy soils in Guizhou, China. Environmental Science: Processes & Impacts, 20: 673–685.
Wahba HM, Lecoq L, Stevenson M, Mansour A, Cappadocia L, Lafrance-Vanasse J, et al. 2016. Structural and biochemical characterization of a copper-binding mutant of the organomercurial lyase MerB: insight into the key role of the active site aspartic acid in Hg-Carbon bond cleavage and metal binding specificity. Biochemistry, 55: 1070–1081.
Yu Q, Szymanowski J, Myneni SCB, and Fein JB. 2014. Characterization of sulfhydryl sites within bacterial cell envelopes using selective site-blocking and potentiometric titrations. Chemical Geology, 373: 50–58.
Yu R-Q, Reinfelder JR, Hines ME, and Barkay T. 2013. Mercury methylation by the methanogen Methanospirillum hungatei. Applied and Environmental Microbiology, 79: 6325–6330.
Yu R-Q, Reinfelder JR, Hines ME, and Barkay T. 2018. Syntrophic pathways for microbial mercury methylation. The ISME Journal, 12(7): 1826–1835.
Zhang DY, Deng CN, and Pan XL. 2013. Excess Ca2+ does not alleviate but increases the toxicity of Hg2+ to photosystem II in Synechocystis sp. (Cyanophyta). Ecotoxicology and Environmental Safety, 97: 160–165.
Zhang H, Feng XB, Larssen T, Qiu GL, and Vogt RD. 2010. In inland China, rice, rather than fish, is the major pathway for methylmercury exposure. Environmental Health Perspectives, 118: 1183–1188.
Zhao LD, Chen HM, Lu X, Lin H, Christensen GK, Pierce EM, et al. 2017. Contrasting effects of dissolved organic matter on mercury methylation by Geobacter sulfurreducens PCA and Desulfovibrio desulfuricans ND132. Environmental Science & Technology, 51: 10468–10475.
Zheng W, and Hintelmann H. 2010. Isotope fractionation of mercury during Its photochemical reduction by low-molecular-weight organic compounds. The Journal of Physical Chemistry A, 114: 4246–4253.
Zhong H, and Wang WX. 2009. Controls of dissolved organic matter and chloride on mercury uptake by a marine diatom. Environmental Science & Technology, 43: 8998–9003.
Zhou J, Riccardi D, Beste A, Smith JC, and Parks JM. 2014. Mercury methylation by HgcA: theory supports carbanion transfer to Hg(II). Inorganic Chemistry, 53: 772–777.
Zhu XF, Zou DH, Huang YH, Cao JM, Sheng GC, and Wang GX. 2015. Physiological responses of Hizikia fusiformis (Phaeophyta) to mercury exposure. Botanica Marina, 58: 93–101.
Information & Authors
Information
Published In

FACETS
Volume 3 • Number 1 • October 2018
Pages: 858 - 879
Editor: Elena P. Ivanova
History
Received: 22 May 2018
Accepted: 30 July 2018
Version of record online: 13 September 2018
Copyright
© 2018 Grégoire and Poulain. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
All conceived and designed the study.
All performed the experiments/collected the data.
All analyzed and interpreted the data.
All contributed resources.
All drafted or revised the manuscript.
Competing Interests
AJP is currently serving as a Subject Editor for FACETS, but was not involved in review or editorial decisions regarding this manuscript.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Daniel S. Grégoire and Alexandre J. Poulain. 2018. Shining light on recent advances in microbial mercury cycling. FACETS.
3(1): 858-879. https://doi.org/10.1139/facets-2018-0015
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Revisiting the mercury cycle in marine sediments: A potential multifaceted role for Desulfobacterota
2. Competition between Dissolved Organic Matter and Freshwater Plankton Control Methylmercury Isotope Fractionation during Uptake and Photochemical Demethylation
3. Mining-impacted rice paddies select for Archaeal methylators and reveal a putative (Archaeal) regulator of mercury methylation
4. The divergent effects of nitrate and ammonium application on mercury methylation, demethylation, and reduction in flooded paddy slurries
5. Plastispheres as hotspots of microbially-driven methylmercury production in paddy soils
6. Mercury and Sulfur Redox Cycling Affect Methylmercury Levels in Rice Paddy Soils across a Contamination Gradient
7. Health risk assessment of methyl mercury from fish consumption in a sample of adult Qatari residents
8. Transcriptional Control of
hgcAB
by an ArsR-Like Regulator in
Pseudodesulfovibrio mercurii
ND132
9. Anthropogenic activity and millennial climate variability affect Holocene mercury deposition of an alpine wetland near the largest mercury mine in China
10. The relationship between zooplankton vertical distribution and the concentration of aqueous Hg in boreal lakes: A comparative field study
11. Microbial communities mediating net methylmercury formation along a trophic gradient in a peatland chronosequence
12. Possible pathways for mercury methylation in oxic marine waters
13. Organo-mercury species in a polluted agricultural flood plain: Combining speciation methods and polymerase chain reaction to investigate pathways of contamination
14. Diffusion of
H
2
S
from anaerobic thiolated ligand biodegradation rapidly generates bioavailable mercury
15. Contrary effects of phytoplankton Chlorella vulgaris and its exudates on mercury methylation by iron- and sulfate-reducing bacteria
16. Experiments revealing the formation of refractory methylmercury pools in natural sediments and soils
17. Highly mercury-resistant strains from different Colombian Amazon ecosystems affected by artisanal gold mining activities
18. Unravelling biogeochemical drivers of methylmercury production in an Arctic fen soil and a bog soil
19. Demethylation─The Other Side of the Mercury Methylation Coin: A Critical Review
20. Mercury in Aquatic Systems of North Patagonia (Argentina): Sources, Processes, and Trophic Transfer
21. Microbial mercury transformations: Molecules, functions and organisms
22. Role of phytoplankton in aquatic mercury speciation and transformations
23. Microbial Communities Mediating Net Methylmercury Formation Along a Trophic Gradient in a Peatland Chronosequence
24. Potential leaching of mercury from stable mercury sulphide
25. Species-specific isotope tracking of mercury uptake and transformations by pico-nanoplankton in an eutrophic lake
26. Stable Isotope Fractionation Reveals Similar Atomic-Level Controls during Aerobic and Anaerobic Microbial Hg Transformation Pathways
27. Mercury methylation and its accumulation in rice and paddy soil in degraded lands: A critical review
28. Expanded Diversity and Phylogeny of mer Genes Broadens Mercury Resistance Paradigms and Reveals an Origin for MerA Among Thermophilic Archaea
29. Mercury methylation by metabolically versatile and cosmopolitan marine bacteria
30. Methylmercury biomagnification in coastal aquatic food webs from western Patagonia and western Antarctic Peninsula
31. Mercury mobility, colloid formation and methylation in a polluted Fluvisol as affected by manure application and flooding–draining cycle
32. Diverse Communities of
hgcAB+
Microorganisms Methylate Mercury in Freshwater Sediments Subjected to Experimental Sulfate Loading
33. Mercury cycling in freshwater systems - An updated conceptual model
34. Mercury biogeochemical cycling: A synthesis of recent scientific advances
35. New Approaches to Understand Mercury in Trees: Radial and Longitudinal Patterns of Mercury in Tree Rings and Genetic Control of Mercury in Maple Sap
36. Biotic formation of methylmercury: A bio–physico–chemical conundrum
37. Overlooked Role of Putative Non-Hg Methylators in Predicting Methylmercury Production in Paddy Soils
38. The microbial mercury link in oligotrophic lakes: Bioaccumulation by picocyanobacteria in natural gradients of dissolved organic matter
39. A Model of Mercury Distribution in Tuna from the Western and Central Pacific Ocean: Influence of Physiology, Ecology and Environmental Factors
