Detection of SARS-CoV-2 in wastewater in Halifax, Nova Scotia, Canada, using four RT-qPCR assays
Abstract
Wastewater-based surveillance methods have been implemented in several countries as a tool for monitoring SARS-CoV-2 at a community scale. A variety of methods have been used for concentrating, extracting, and detecting the virus, with no clear consensus on the most effective approach. In this note, we report preliminary findings from a study that is tracking SARS-CoV-2 in wastewater in Halifax, Nova Scotia, with a specific focus on the use of four reverse transcriptase quantitative PCR (RT-qPCR) assays for detecting the virus in wastewater. We were able to detect the virus in wastewater samples during the initial rise of cases in the Halifax region in early November 2020. Levels of the targeted SARS-CoV-2 gene fragments increased and fell in response to reported cases of COVID-19. The CDC N1 and E RT-qPCR assays demonstrated greater relative sensitivity than the CDC N2 and N3 assays for detection of SARS-CoV-2 in raw sewage samples.
Introduction
The ongoing pandemic associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in 115 653 459 cases and 2 571 823 deaths worldwide as of 6 March 2021 (WHO 2021). To date, Canada has reported 884 086 cases and 22 213 deaths (Government of Canada 2021). The development of effective testing and surveillance methods is key to managing the impacts of the pandemic as vaccines are being developed and distributed. Wastewater-based surveillance, which involves detection of SARS-CoV-2 RNA fragments in community sewage matrices, has emerged as a potentially important surveillance and early warning tool. Initial studies conducted in the Netherlands (Medema et al. 2020) and the United States (Peccia et al. 2020; Wu et al. 2020) demonstrated how this method could provide advanced notice of increases in community infections. Additional studies assessing the use of wastewater-based SARS-CoV-2 surveillance in several other regions have since been reported. Examples include reports by Ahmed et al. 2020 (Australia), Randazzo et al. 2020 (Spain), Kumar et al. 2020 (India), Hata et al. 2021 (Japan), and D’Aoust et al. 2021 (Ottawa, Canada). The majority of studies have utilized RT-qPCR for detection of the fragments of the viral genome, with a variety of methods employed for concentration and extraction of the virus from wastewater samples (Philo et al. 2021). The RT-qPCR assays, which have been used for detection and quantification of the virus in wastewater, are primarily those that have been developed for clinical testing, such as the US Centers for Disease Control and Prevention (CDC) N1, N2, N3 assays targeting different regions of the nucleocapsid gene, and the E gene assay, which targets the virus envelope region (Medema et al. 2020).
The province of Nova Scotia, Canada, has reported relatively few cases (1657) and deaths (65), which is likely due to a combination of factors including low population densities, geographic location, and the quick implementation of public health measures such as mandatory mask usage, travel restrictions, and gathering restrictions. Nova Scotia reported its first presumptive cases of COVID-19 on 15 March 2020 and saw a total of 1063 cases and 63 deaths during the first wave of the pandemic (to 1 July 2020) (Government of Nova Scotia 2021). Case numbers remained low throughout the summer and fall months, but the province started to experience a second wave of infections in the Halifax region in November 2020. Lockdown measures were immediately implemented and were effective in reducing infection rates to pre-second wave levels by early January 2021. We initiated a study to monitor SARS-CoV-2 in wastewater in the capital city of Halifax, Nova Scotia, in early fall of 2020. Here, we report on the findings from the first five months of this monitoring program, which documents the performance of this method in detecting the rise and decline of cases during the second wave of SARS-CoV-2 infections in the Halifax region. We focus on the use of four different RT-qPCR assays for the detection of the virus in wastewater.
Methods
Samples were collected and analyzed on a weekly basis from the Halifax Wastewater Treatment Facility (Fig. 1) starting the first week of October 2020. The samples were 24 h composite samples obtained from the influent to the wastewater plant using autosamplers. The samples were transported to Dalhousie University at a temperature of 4 °C the day of collection and either processed immediately or frozen until the analysis could be completed. The Halifax Wastewater Treatment Facility services peninsular Halifax and portions of Bedford (Halifax Water 2021), which would have a population of approximately 120 000.
Fig. 1.
The extraction and detection of SARS-CoV-2 in wastewater samples followed the methodology presented in Medema et al. (2020), with minor adaptations. Briefly, the procedure employs centrifugal ultrafiltration for viral concentration. We used Centricon® Plus-70 centrifugal ultrafilters (Sigma-Aldrich, Oakville, ON, Canada) with a molecular weight cut-off of 10 kDa, 200 mL sample volumes, and a centrifuge speed of 4000g for 30 min. RNA was extracted from the sample concentrates (ranging from 0.80g to 1.66 g) using Qiagen RNeasy Power Microbiome Kit (Qiagen Inc., Toronto, Ontario, Canada) according to the manufacturer’s instructions. RNA was also extracted from 50 mL of Milli-Q water (Sigma-Aldrich, Oakville, ON, Canada) that served as a negative control to test if cross-contamination had occurred during the whole RNA extraction process.
The four RT-qPCR assays we used were the CDC N1, N2, N3 assays and the E assay (Corman et al. 2020). The analysis was carried out on a Bio-Rad CFX96 TouchTM Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). Standard curves for the N1, N2, and N3 assays were constructed using the undiluted (1 × 106 copies/reaction) and 10-fold diluted standards ranging in concentration from 1 × 105 to 1 × 101 copies/reaction of 2019-nCOV_N_Positive Control Plasmid DNA from IDT (Integrated DNA Technologies, Inc., Coralville, IA, USA). The standard curve for the E assay was built using the undiluted (5.3 × 105 copies/reaction) and 10-fold diluted (ranging from 5.3 × 104 to 5.3 × 101 copies/reaction) suspensions of RNA extracted from the ATCC VR-1986HK heat-inactivated 2019 novel coronavirus (ATCC, Manassas, VA, USA). Two technical replicates were run for all standards, samples, and nontemplate controls and the difference of the threshold cycle (Ct) value between the replicates was always >0.5. The N1 assay efficiency was 100.7%, with an R2 value of 0.995. The N2 assay efficiency was 107.8%, with an R2 value of 0.998. The N3 assay efficiency was 101.9%, with an R2 value of 0.995. For the N1–N3 assays, the limit of detection (LOD) was the same as the limit of quantification (LOQ), which was determined to be 10 copies/reaction. The E assay efficiency was 99.9%, with an R2 value of 0.996. Similar to the N assays, the LOD equalled the LOQ and was determined to be 52.5 copies/reaction. In each assay, the absence of RT-qPCR inhibitors was confirmed by comparison of Ct values obtained for aliquots of undiluted and 10-fold diluted RNA extract from samples, which had been spiked with known concentrations of the positive controls for the N and E assays.
We evaluated the recovery efficiency by spiking wastewater samples with the ATCC VR-1986HK heat-inactivated 2019 novel coronavirus and found it to be 16%. Medema et al. (2020) evaluated the recovery efficiency of this methodology by examining the recovery of F specific RNA phages, reporting a recovery efficiency of 73%. It would be expected that the recovery efficiency of an enveloped virus such as SARS-CoV-2 would be lower than for nonenveloped F specific RNA phages (Lu et al. 2020).
The presence and levels of SARS-CoV-2 genetic targets were reported as gene copies per millilitre in wastewater and compared to available COVID-19 case data published by the Province of Nova Scotia (Government of Nova Scotia 2021). In Nova Scotia, COVID-19 cases are reported for each of the four Health Management Zones. The city of Halifax is located within the Central Health Management Zone (CHMZ), which also includes several other communities and has a total population of approximately 424 000 (Fig. 1).
Results and discussion
Throughout October 2020, Nova Scotia was reporting less than three cases per day in the CHMZ and the active case numbers were less than ten (Fig. 2). During this time, we did not detect any of the four SARS-CoV-2 genetic targets in samples collected from the influent of the Halifax Wastewater Treatment Facility. The first wastewater sample that tested positive for SARS-CoV-2 was collected on 12 November 2020, with the N1, N3, and E assays producing positive signals. Concentrations of the N1 and N3 gene fragments were 1.6 and 1.5 copies/mL, respectively, and the concentration of the E gene fragment was 1.2 copies/mL. The active cases in the CHMZ had increased to 19 by this date. During the next three weeks, all four assays were positive in influent wastewater at the treatment facility, with SARS-CoV-2 levels increasing each week. Maximum concentrations of the N1, N2, N3, and E gene fragments were 24.5, 23.4, 25.6, and 23.2 copies/mL, respectively (Fig. 2), which coincided with active cases in the CHMZ having increased to a maximum of 138 during that week. The Nova Scotia government implemented a suite of lockdown measures during the last week of November 2020, which resulted in a progressive reduction in new and active cases throughout the month of December 2020. SARS-CoV-2 gene fragment levels decreased during this time as well, with only the N1 and E assays producing positive signals during the week of 16 December 2020. None of the assays were positive from 23 December 2020 to 6 January 2021. On 13 January 2021, the N1 and E assays produced weak positive signals, corresponding with a slight increase in active cases, which was likely associated with university students returning to Halifax after the holiday break. Case numbers continued to decline after 13 January 2021 and the four targets were undetected in wastewater until mid-February, 2021. The N1 assay produced a weak positive signal on 17 February, and again on 24 February and 3 March 2021. The E assay was also positive on 24 February and 3 March. These positive signals coincided with a small increase in active cases in the CHMZ, which peaked at 39 cases on 27 February 2021 and then subsequently declined. The N2 and N3 assays did not produce positive signals during this time period.
Fig. 2.
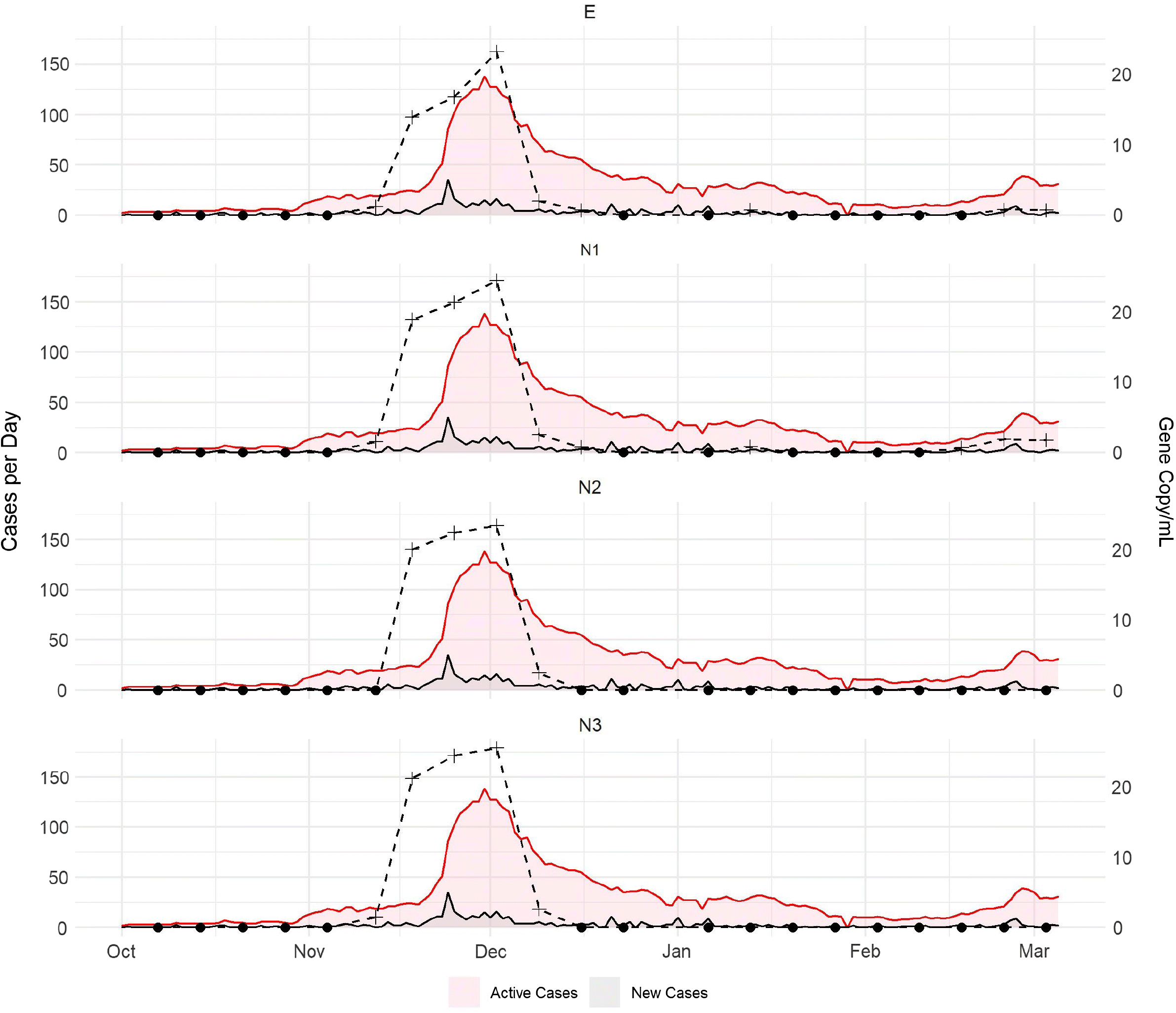
These results have demonstrated that wastewater-based surveillance could be a useful tool for monitoring SARS-CoV-2 in regions with a low prevalence of the virus. In general, we were able to detect the virus in influent wastewater when active case numbers increased above 20 within the CHMZ. The number of active cases within the sewershed for the Halifax Wastewater Treatment Facility would likely have been lower than this, as it only captures wastewater from a portion of the total population of CHMZ, which may underpin the sensitivity of this surveillance strategy. Positive marker signals were also present approximately one week prior to a steep rise in new cases in the CHMZ in mid-November (Fig. 1), which illustrates the potential for this tool to be used for early warning of increasing cases. Of particular note was the difference in detection frequency of the four RT-qPCR assays, with the N1 and E assays displaying greater sensitivity to increases in active cases. There are only a few studies that specifically compared the detection frequency of the different SARS-CoV-2 assays for wastewater and they have produced conflicting results. Medema et al. (2020) also found that the N1 and N3 assays were positive prior to the N2 assay. In contrast, Gonzalez et al. (2020) reported a higher frequency of detection with the N2 target. However, they noted that their RT-qPCR detection limit for the N1 assay was higher than the N2 and N3 assays. D’Aoust et al. (2021) evaluated the N1, N2, N3, and E assays and found better detection and lower variance with the N1 and N2 assays. Further research needs to be conducted to better understand the relative sensitivity of the assays currently in use for wastewater-based SARS-CoV-2 surveillance. However, these findings demonstrate the benefits of using multiple gene targets for wastewater-based SARS-CoV-2 surveillance to provide increased confidence in positive or negative signals in regions with low virus prevalence. Future studies could also include the quantification of pepper mild mottle virus or HF183 Bacteroides 16S ribosomal RNA as an indicator of variability in the sewage system (e.g., dilution due to inflow/infiltration) or sampling procedures (D’Aoust et al. 2021) to help further assess assay differences.
Conclusions
This study demonstrated that wastewater-based surveillance was successful in tracking the rise and decline of the second wave of SARS-CoV-2 infections in the Halifax region of Nova Scotia during the fall of 2020. Of the four RT-qPCR assays used, the N1 and E assays were found to be more sensitive to rising infection rates.
Acknowledgements
This research has been funded by the Natural Sciences and Engineering Research Council of Canada (NSERC Alliance Grant ALLRP 554503 – 20) and Research Nova Scotia. We would like to acknowledge Halifax Water for assistance with sample collection and LuminUltra Technologies Ltd.
References
Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, et al. 2020. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment, 728: 138764.
Corman, VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RTPCR. European Surveillance, 25(3): 2000045.
D’Aoust PM, Mercier E, Montpetit D, Jia J, Alexandrov I, Neault N, et al. 2021. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Research, 188: 116560.
Gonzalez R, Curtis K, Bivins A, Bibby K, Weir M, Yetka K, et al. 2020. COVID-19 surveillance in Southeaster Virginia using wastewater-based epidemiology. Water Research, 186: 116296.
Government of Canada. 2021. Coronavirus disease (COVID-19) outbreak update. [online]: Available from canada.ca/en/public-health/services/diseases/coronavirus-disease-covid-19.html.
Government of Nova Scotia. 2021. Coronavirus (COVID-19) case data. [online]: Available from novascotia.ca/coronavirus/data/.
Halifax Water. 2021. Maps and apps: Where does my wastewater go. [online]: Available from halifaxwater.ca/maps-apps.
Hata A, Hara-Yamamura H, Meuchi Y, Imai S, and Honda R. 2021. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Science of the Total Environment, 758: 143578.
Kumar M, Patel AK, Shah AV, Raval J, Rajpara N, Joshi M, and Joshi CG. 2020. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Science of the Total Environment, 709: 141326.
Lu D, Huang Z, Luo J, Zhang X, and Sha S. 2020. Primary concentration: The critical step in implementing the wastewater based epidemiology for the COVID 19 pandemic. A mini-review. Science of the Total Environment, 747: 141245.
Medema G, Heijnen L, Elsinga G, Italiaander R, and Brouwer A. 2020. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the pidemic in The Netherlands. Environmental Science and Technology Letters, 7: 511–516.
Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, et al. 2020. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology, 38: 1164–1167.
Philo SE, Keim EK, Swanstrom R, Ong AQW, Burnor EA, Kossik AL, et al. 2021. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Science of the Total Environment, 706: 194215.
Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, and Sánchez G. 2020. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Research, 181: 115942.
World Health Organization (WHO). 2021. Coronavirus disease (Covid-19) dashboard. [online]: Available from covid19.who.int/.
Wu F, Zhang J, Xiao A, Gu X, Lee WL, Armas F, et al. 2020. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems, 5: 20051540.
Information & Authors
Information
Published In

FACETS
Volume 6 • Number 1 • January 2021
Pages: 959 - 965
Editor: Yang Liu
History
Received: 8 March 2021
Accepted: 23 April 2021
Version of record online: 10 June 2021
Copyright
© 2021 Huang et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
YH, AP, CS, GG, AS, and RJ conceived and designed the study.
YH, AP, CS, and EH performed the experiments/collected the data.
YH, LJ, LTH, and RJ analyzed and interpreted the data.
LTH, GG, AS, and RJ contributed resources.
YH, LJ, and RJ drafted or revised the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Yannan Huang, Lindsay Johnston, Ana Parra, Crystal Sweeney, Emalie Hayes, Lisbeth Truelstrup Hansen, Graham Gagnon, Amina Stoddart, and Rob Jamieson. 2021. Detection of SARS-CoV-2 in wastewater in Halifax, Nova Scotia, Canada, using four RT-qPCR assays. FACETS.
6(): 959-965. https://doi.org/10.1139/facets-2021-0026
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Correlation between SARS-CoV-2 RNA concentration in wastewater and COVID-19 cases in community: A systematic review and meta-analysis
2. Artificial neural network-based estimation of COVID-19 case numbers and effective reproduction rate using wastewater-based epidemiology
3. SARS-CoV-2 in wastewater: From detection to evaluation
