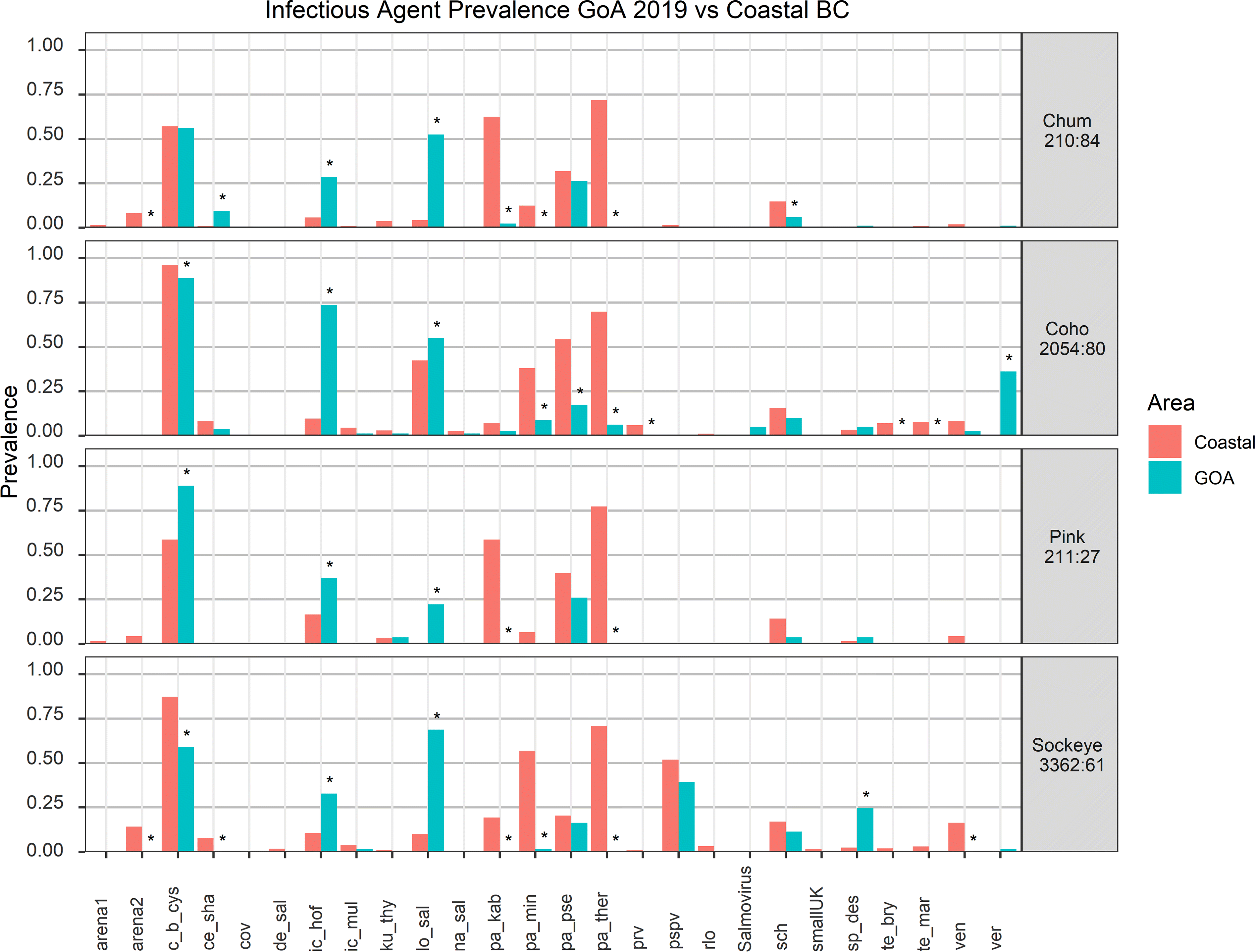
Prey availability, temperature-related factors, and infectious agent profile correlate with differential gene expression of salmon in the GoA
To investigate stressors of salmon in the GoA, we compared the expression of all genes from all biomarker panels across all individuals of the same species. First, we visualized gene expression using heatmaps, also displaying pathogen detections as well as co-varying metadata (
Fig. 3). Hierarchical clustering of gene expression allowed us to identify clusters of salmon showing similar expression patterns (
Fig. 3). In chum, clusters four and five showed markedly reduced overall gene expression that was associated with elevated RIB and lower biomass of hydromedusa at capture location, the primary prey of chum salmon (
Somov et al. 2019), as well as lower levels of dissolved oxygen (
Fig. 3;
Supplementary Material, Fig. 5). Further, temperature at site of capture was also significantly associated with overall gene expression, with warmer temperatures correlating to higher gene expression (
Fig. 3;
Supplementary Material, Fig. 5). Similarly, in sockeye, elevated temperature, and prey availability (e.g., small zooplankton) was associated with a global increase in gene expression (
Fig. 3;
Supplementary Material, Fig. 6). Condition factor K was significantly covaried between clusters in sockeye (
Fig. 3;
Supplementary Material, Fig. 6). Coho salmon showed no large-scale changes in gene expression and clusters differed primarily in the response of individual biomarkers to RIB and prey availability (
Fig. 3;
Supplementary Material, Fig. 7). Pink salmon also showed large-scale changes to gene expression associated with RIB, prey availability, and temperature, but interestingly high values of these factors were associated with reduced global gene expression rather than an increase as had been seen in chum and sockeye salmon (
Fig. 3;
Supplementary Material, Fig. 8).
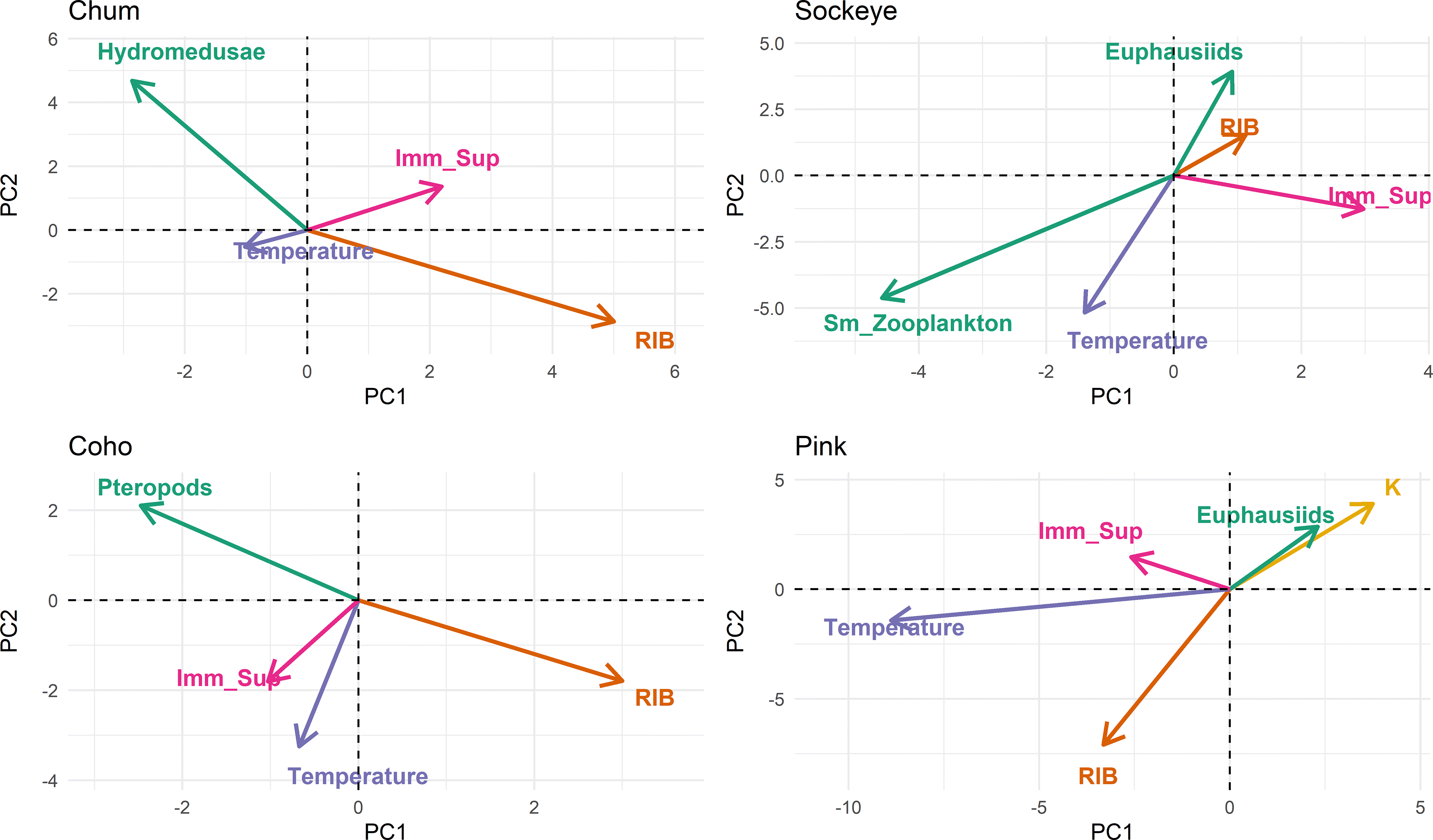
Next, we performed a PCA to ordinate gene expression profiles of individual salmon. We focused on the first four principal components to identify dominant biomarker panels driving differential gene expression. We then tested observational data on salmon health and condition as well as oceanographic data for correlations with principal components and plotted significantly correlated data scaled and directional on the ordination plots to depict the direction of correlation. In the last step, we queried what genes showed changes in expression correlated with the superimposed data by using a Euclidean distance-based approach or plotted a vector summarizing of all genes of a biomarker panel, respectively (
Fig. 4;
Supplementary Material, Fig. 9). By visualizing the hierarchical clusters identified earlier, this allowed us to identify the environmental factors and pathogens associated with the differential gene expression providing a population scale overview of stressors of overwintering salmon (
Fig. 4;
Supplementary Material, Fig. 9).
Differential gene expression in chum salmon was primarily driven by variations in biomarkers for inflammation (MMP13, NAPEPLD2, TXN, GILT), immune stimulation (SAA, CD83, IFNa), mortality-related (C7, P_RAS), viral disease development (VDD) biomarker panels (HERC6, IFIT5, IFI44, VAR1), followed by imminent mortality and hypoxia (TAGLN3, CDKN1B;
Fig. 4a;
Supplementary Material, Fig. 9a). Along PC1, these factors explained 36% of the variation in gene expression. Chum clusters four and five showed lower gene expression across all biomarker panels and clustered on the positive end of PC1 (
Figs. 3 and
4a;
Supplementary Material, Fig. 9a). Inflammation (MMP13, NAPEPLD2, TXN) and immune stimulation biomarkers (SAA) contributed the negatively to PC2 (10.8% explanatory power), while hypoxia and imminent mortality biomarkers (CDKN1B) contributed positively (
Figs. 3 and
4a;
Supplementary Material, Fig. 9a). RIB as well as nematode prevalence was correlated with lower overall gene expression in individuals of cluster four and five, but positively associated with inflammation (MMP13, NAPEPLD2), immune stimulation (SAA), and VDD (HERC6, IFIT5) markers on PC2. Conversely, pathogens
P. pseudobranchicola (pa_pse) and
S. destruens (sp_des) were negatively associated with these immune response markers along PC2 (
Fig. 4a;
Supplementary Material, Fig. 9a). Biomass of hydromedusae (Medu) and other the prey of chum (small zooplankton: Zoo_S) was positively correlated with global upregulated gene expression along PC1 and lower expression of the immune response markers associated with PC2 (
Somov et al. 2019), while being directly opposed to RIB and nematode prevalence across PC1 and PC2 (
Fig. 4a;
Supplementary Material, Fig. 9a). Principal components three and four (explaining 10% and 6.2%, respectively) were driven by the same genes driving PC1 and PC2; however, inflammation and VDD markers were opposing each other along PC3, with inflammation driven by individuals of cluster four, that showed enlarged gallbladders (a sign of prolonged low stomach fullness) and larger size (Mass) and smaller individuals at higher temperature associated with VDD expression (
Fig. 4b;
Supplementary Material, Fig. 9b). PC4 was driven by opposing trends of immune stimulation and hypoxia biomarkers, primarily associated with zooplankton (euphausiids and medium size zooplankton;
Figs. 3 and
4b;
Supplementary Material, Fig. 9a).
Sockeye showed similar patterns to chum salmon with two clusters (one and four) showing reduced overall gene expression associated with the positive end of PC1 (43.8%). The primary drivers associated with these global expression changes were the biomarker panels immune stimulation (B2M, HEP, IGMs, CD83, SAA), inflammation (IL_17D, ES1), mortality related (SCG2, RPL6), VDD (HERC6, DEXH, MX, IFI), and a group of hypoxia genes (RRl, CLASPIN, KIF15, COX6B, RRM2) (
Fig. 4c;
Supplementary Material, Fig. 9c). These hypoxia genes were also major contributors to PC2 (13.8%) opposed by the general stress marker JUN_F3 (
Fig. 4c;
Supplementary Material, Fig. 9c). Globally lowered gene expression in sockeye clusters one and four was associated with lower abundance of small zooplankton (Zoo_S), pteropods (Ptero), and hydromedusae (Medu) along PC1, and to a lesser degree lower temperature at site of capture (
Fig. 4c;
Supplementary Material, Fig. 9c). Euphausiids (Euphaus) that were identified as the primary prey of sockeye, were correlated with the positive end of PC2, opposed to temperature, and showed increased expression of inflammation and immune stimulation markers, but showed weaker association with gene expression than other prey groups (
Fig. 4c;
Supplementary Material, Fig. 9c). The prevalence of the gill parasite
Loma spp. (lo_sal) was associated with expression of inflammation and immune stimulation biomarkers along PC1 and PC2. Principal component three (7.7% exploratory power) saw a strong correlation of immune stimulation (SAA, IFNa, IGM) and inflammation biomarkers (IL_17D, MMP24, MMP13) with the parasites
I. hoferi and
P.
pseudobranchicola, whereas inflammation (ES1, EPD) and imminent mortality markers (TAGLN3, RGS21) were associated with nematode prevalence (
Fig. 4d;
Supplementary Material, Fig. 9d). Fish with higher caloric content and better condition factor (K) were also associated with lower temperatures at site of capture and lower prevalence of pathogens (ic_hof, pa_pse) (
Fig. 4d;
Supplementary Material, Fig. 9d).
Differential gene expression in coho salmon showed a nuanced response of biomarker panels along the first two principal components where inflammation (MMP13, IL_11, NAPEPLD2, IL_17D), general stress (JUN_F3), immune stimulation (IL_1b, HEP, SAA, IFNa), and VDD (HERC6, GAL3) associated positively with RIB and fish of cluster four on the positive end of PC1 (21.2%;
Fig. 4e;
Supplementary Material, Fig. 9e). RIB was inversely related to the biomass of pteropods (ptero) that were the preferred prey of coho salmon in GoA in 2019 (
Somov et al. 2019), with fish from cluster four experiencing the lowest pteropod biomass (
Fig. 4e;
Supplementary Material, Fig. 9e). Hypoxia biomarkers (COX6B, RRM2, CDKN1B) were correlated with the prevalence of the gill parasite
Loma sp. (lo_sal) along PC1 and PC2 (17.5%) (
Fig. 4e;
Supplementary Material, Fig. 9e), specifically amongst individuals of clusters two, three, and five. Principal components three and four (12.4% and 7.1%, respectively) showed a global increase in expression that was associated with the size of fish (Mass) of individuals in cluster four, as well as an increased expression of VDD biomarkers related to
P.
pseudobranchicola (pa_pse) load (
Fig. 4f;
Supplementary Material, Fig. 9f).
Pink salmon of cluster one showed reduced global gene expression compared to other clusters, grouped along the negative spectrum of PC1 (64.6 %) and were associated with higher temperatures, higher RIB, and higher biomass of prey species (
Fig. 4g;
Supplementary Material, Fig. 9g). On the positive spectrum of PC1, clusters two, three, and four were associated with increased expression imminent mortality markers (CDKN1B, CBEBP, GPX) but were differentiated along PC2 (10.2%) with expression of hypoxia (COX_6B, RRI, GPX), and inflammation biomarkers (NAPEPLD2, IL_17D) were associated with cluster four, while clusters two and three showed increase expression of VDD (TRIM, GAL3, MX, VAR1, IFI) and immune stimulation markers (SAA, IGMs, HEP, CD83) that were associated with increased RIB, number of infectious agents, as well as the prevalence of the parasites
S. destruens (sp_des) and
P. psudobranchiola (pa_pse) (
Fig. 4g;
Supplementary Material, Fig. 9g). Principal component three (explaining 5.6%) showed elevated expression of VDD (GAL3, Mx, IFI, HERC6), immune stimulation (SAA, IL_15), and general stress genes (HSP90) along the positive end of PC3, which was correlated with larger individuals (Mass;
Fig. 4h;
Supplementary Material, Fig. 9h). Higher biomass of Euphausiids (Euphaus) along PC4 (4.1%) correlated with VDD expression and inflammation (EPD).
To highlight overlying trends of pathogens and environmental factors such as prey biomass and temperature, we plotted the biomass of primary prey species in relation to ocean temperature and RIB across the first two principal components of gene expression (
Fig. 5;
Supplementary Material, Fig.10). Since global depression of immune response genes (immune stimulation, inflammation, and viral disease development) effectively equals immunosuppression, we created the inverse vector of gene expression of said biomarker panels to depict this suppressed immune status. Indeed, immunosuppression showed an inverse relationship with the biomass of the primary prey species as well as a direct correlation with RIB in all species (
Fig. 5). In chum and pink salmon this trend dominated gene expression along PC1 (
Fig. 5). Coho showed a strong inverse correlation between primary prey biomass and RIB, but immunosuppression was only weakly associated with them along PC2, suggesting that large-scale changes in gene expression resulting in immunosuppression are subordinate to other factors relating to RIB (
Fig. 5). In sockeye, gene expression patterns were more strongly associated with small zooplankton, rather than the primary stomach content which was Euphausiids (
Fig. 5). Accordingly, lower biomass of small zooplankton was associated with immunosuppression and elevated RIB in sockeye along PC1 (
Fig. 5). Coho and pink salmon that were primarily caught along the southern border of the distribution area and experienced the highest ocean temperatures and showed a strong correlation of immunosuppression and RIB with increased temperature (
Fig. 5).
Infectious agent profiles correlated with gene response to viral and gill infections and stock of origin in coho
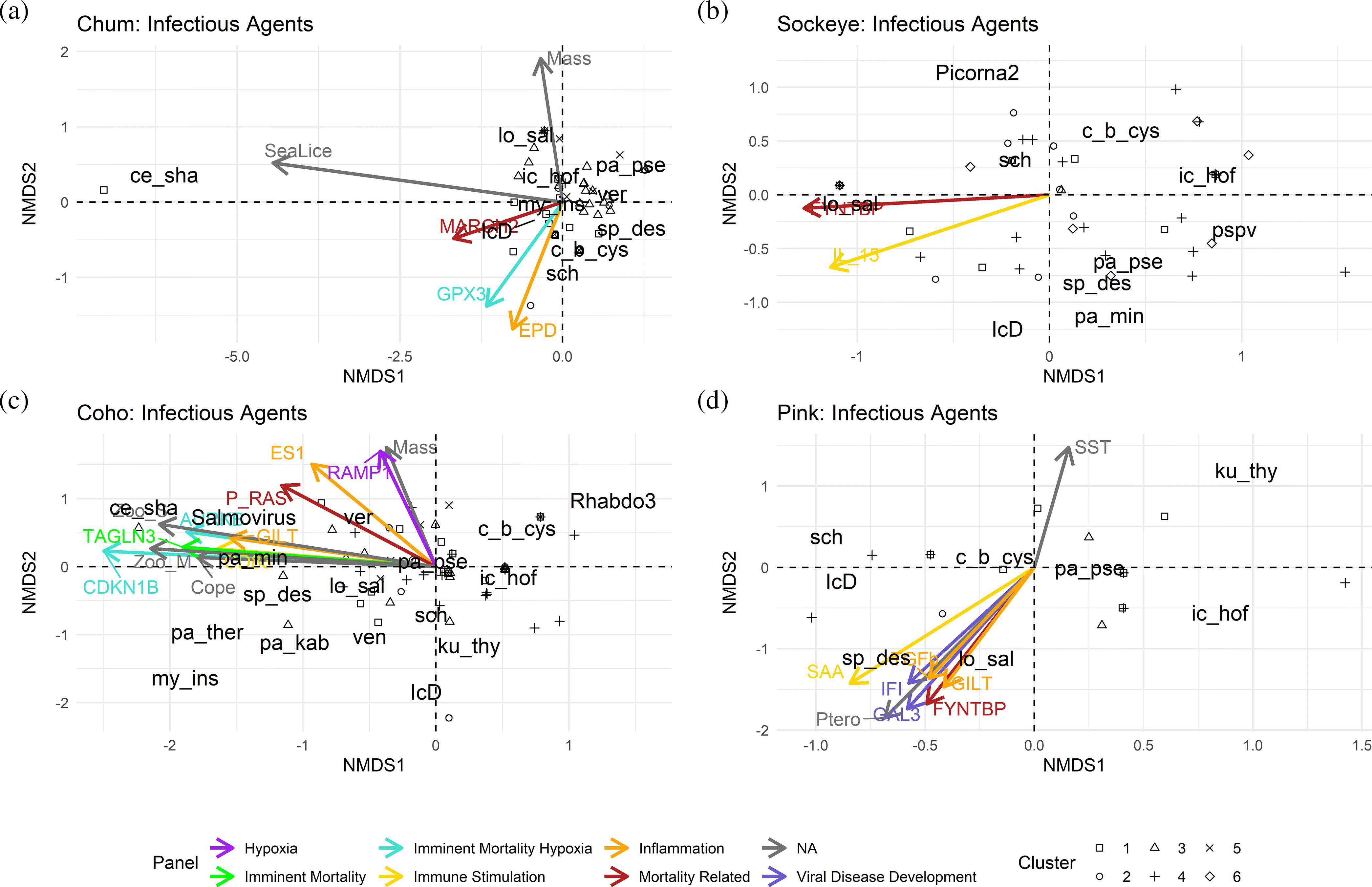
To determine if infectious agent profiles were associated with environmental factors and gene expression, we visualized the gene expression data in rank order-based NMDS-ordinated pathogen profiles of individuals by species (
Fig. 6).
Differences in chum infectious agent profiles were primarily driven by
C. shasta (ce_sha) with minor opposing contributions of
P. pseudobranchicola (pa_pse) and
S. destruens (sp_des) along NMDS1 (
Fig. 6a).
C. shasta was only found in individuals of gene expression cluster one and was associated with the expression of a mortality-related biomarker (MARCH2) (
Fig. 6a). NMDS2 differentiation was driven by
Loma sp. (lo_sal) and
P. pseudobranchicola (pa_pse) on the positive end of NMDS2 that were correlated with larger individuals (Mass;
Fig. 6a). Smaller individuals on the negative end of NMDS2 were associated with
Ca. S.
salmonis (sch) and Ca. B.
cysticola (c_b_cys), as well as the expression of imminent mortality/hypoxia (GPX3) and inflammation (EPD) biomarkers.
Sockeye infectious agent profiles differed primarily by the opposing trends of
Loma sp. (lo_sal) against PSPV and
I. hoferi (ic_hof) along NMDS1 with mortality related (FYNTBP) and immune stimulation biomarkers (IL_15) associated with
Loma sp. (
Fig. 6b). Differences across NMDS2 were driven by Putative-picornavirus (Picorna2),
Ichthyobodo sp. (IcD), and
P. minibicornis (pa_min) (
Fig. 6b).
Stock of origin was significantly associated with pathogen profile variation in coho salmon. Accordingly, the pathogen profiles were primarily differentiated by rare and stock-specific pathogens such as
C. shasta (ce_sha), Salmovirus,
P. minibicornis (pa_min),
P. theridion (pa_ther), and
M. insidiosus (my_ins) along the negative end of NMDS1, present in only a few individuals each; the latter three pathogens were only found in fish originating from within the contiguous United States (
Fig. 6c). Correlating gene expression was seen in genes from biomarker panels imminent mortality and hypoxia (CDKN1B, TAGLN3, AURKB), inflammation (GILT, ES1), immune stimulation (CD83), mortality-related signature (P_RAS), as well as the prevalence of medium-sized and small zooplankton (Zoo_S/M) (
Fig. 6c). Hypoxia gene expression (RAMP1) was correlated with large individuals along NMDS2, while small individuals were associated with
Ichthyobodo sp. (IcD) (
Fig. 6c).
Infectious agent profiles in pink salmon differed primarily in the presence of Ca. S
. salmonis (sch),
S. destruens (sp_des), and
Ichthyobodo sp. (IcD) opposed by
I. hoferi (ic_hof) and
K. thyrsites (ku_thy) along NMDS1 (
Fig. 6d). Immune stimulation (SAA), inflammation (TGFb, GILT), VDD (IFI, GAL3) and mortality related biomarkers (FYNTBP) were correlated with the gill pathogen
Loma sp. (lo_sal) and
S. destruens (sp_des) and at higher abundance of pteropods (Ptero) and lower sea surface temperature (SST;
Fig. 6d).