Introduction
Piscine orthoreovirus (PRV) is a common virus of salmon that is endemic (i.e, has maintained constant presence and/or usual prevalence without a need for external introduction) to North America, Europe, South America, and Japan (
Polinski et al. 2020). Infections typically manifest as high-load blood infections where red blood cells are the primary target for PRV replication (
Wessel et al. 2015). In many instances, fish do not clear infections once infected, and the virus can persist in infected populations at relatively high loads for over a year (
Garver et al. 2016). The virus has also been refractory to
in vitro culture (
Pham et al. 2020), thus requiring molecular methods for diagnosis and load estimations (
Palacios et al. 2010;
Polinski et al. 2019).
Phylogenetic analysis of PRV has identified three major genotypes: PRV-1, PRV-2, and PRV-3. There are also at least two subgenotypes for PRV-1 (PRV-1a and 1b) and PRV-3 (PRV-3a and 3b) (
Siah et al. 2020;
Sørensen et al. 2020;
Godoy et al. 2021). Genotype occurrence roughly corresponds to discrete hosts and/or regions (
Polinski et al. 2020). PRV-1 is found predominately in Atlantic, Chinook, and coho salmon of Europe and the Americas, PRV-2 has only been detected in coho salmon of Japan, and PRV-3 is primarily found in rainbow trout and brown trout in Europe and rainbow trout and coho salmon in Chile (
Kibenge et al. 2013;
Takano et al. 2016;
Cartagena et al. 2020). Separate diagnostic assays that have been developed to detect PRV-1, PRV2, and PRV-3 are not effective for cross-genotype detections. Thus, identification of multiple PRV genotypes within a single fish requires multiple assays to be run, or a recently developed Pan-PRV assay must be utilized (
Zhao et al. 2021).
Phylogenetic characterization of PRV-1 has identified regional differentiation within the PRV-1a and PRV-1b subgenotypes, where both subgenotypes co-occur in Norway and Chile, whereas only PRV-1a has been identified in the northwestern Atlantic and northeastern Pacific (
Siah et al. 2020). Indeed, PRV-1a endemic to the northeastern Pacific can be differentiated from PRV-1a found in the Atlantic through phylogenetic comparison, either by whole-genome sequencing or individually comparing the S1 genomic segment of the virus (
Kibenge et al. 2019;
Siah et al. 2020;
Godoy et al. 2021). Thus, putative introductions of nonendemic forms of PRV to the northeastern Pacific (either genotype or subgenotype) should be readily identifiable through whole-genome or S1 comparisons.
PRV has sometimes been associated with disease for which variations in pathogenicity can partially be explained by genotype/subgenotype variations (
Polinski et al. 2020;
Wessel et al. 2020). In Norway, challenge trials using PRV-1b have demonstrated a causal link between PRV-1b and a disease known as heart and skeletal muscle inflammation (HSMI) in farmed Atlantic salmon (
Wessel et al. 2017). Interestingly, challenge trials using PRV-1a from the Atlantic have demonstrated less pathogenicity compared with PRV-1b (
Wessel et al. 2020), and numerous challenge trials involving PRV-1a in the northeastern Pacific have demonstrated low virulence to both Atlantic and Pacific salmon (
Garver et al. 2015,
2016;
Polinski et al. 2019,
2021b;
Zhang et al. 2019;
Purcell et al. 2020). In Japan, PRV-2 is considered a driver of an anemic condition of farmed coho salmon known as erythrocytic inclusion body syndrome (
Takano et al. 2016), and PRV-3 causes an HSMI-like condition of rainbow trout in Europe (
Vendramin et al. 2019) and jaundice/anemia syndrome in farmed coho salmon in Chile (
Cartagena et al. 2020). There may also be an association between PRV-1a and heart inflammation and/or anemia in some instances (
Cicco et al. 2017,
2018). Introductions of PRV genotypes/subgenotypes into nonendemic regions could therefore have potential health implications for resident salmon populations.
In the northeastern Pacific, the occurrence of PRV-1a is well documented in ocean stocks of salmon (
Polinski et al. 2020). However, occurrence of PRV-1a (or any other genotype/subgenotype) in freshwater juvenile anadromous salmon within the region is far less clear. It is apparent that nearly all net-pen farmed salmon in British Columbia, Canada, become infected with PRV-1 within the first 18 months at sea (
Polinski et al. 2022), and cumulative data from multiple wild Pacific salmon surveys have identified an overall regional prevalence in ocean-caught Chinook and coho salmon between 2013 and 2019 of approximately 6% (
Polinski et al. 2020). Historically, there has been at least some evidence that PRV-1 has occurred in commercial Canadian salmon hatcheries rearing juvenile fish (
Marty et al. 2015;
Garver et al. 2016), although distribution and temporal changes in prevalence within freshwater facilities are unknown. There have been no reports for PRV-2 or PRV-3 in the northeastern Pacific, although we are unaware of any studies that have actively targeted either of these genotypes for diagnostics.
In this study, our aims were to (1) define the current regional prevalence of PRV-1 in freshwater anadromous salmon hatchery facilities of western Canada, (2) identify whether alternative genotypes/subgenotypes (e.g., PRV-1b, PRV-2, or PRV-3) are currently occurring within freshwater populations in the region, and (3) confirm interlaboratory agreement for PRV testing between regional laboratories.
Materials and methods
Sample collection
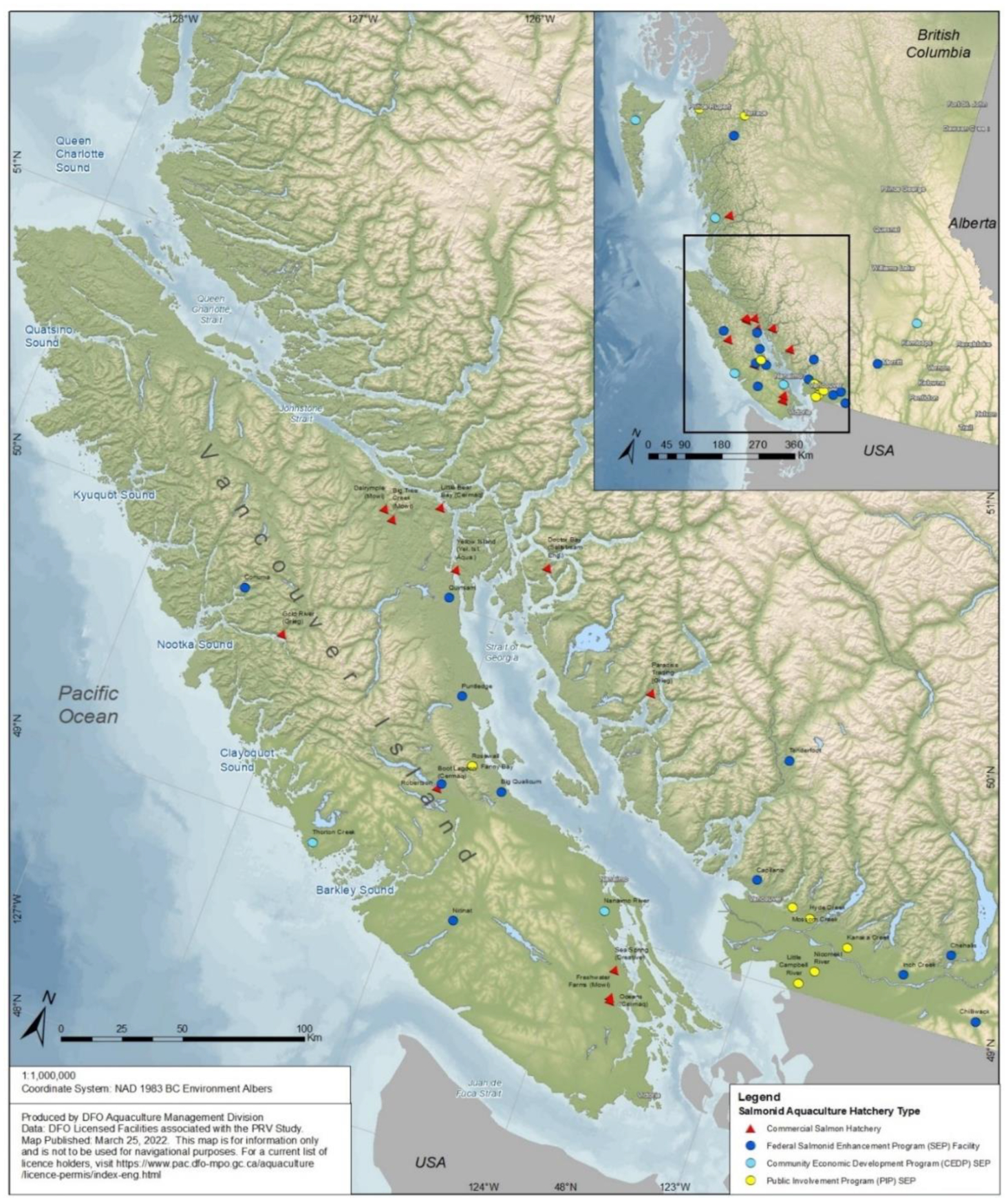
Freshwater anadromous salmon hatcheries in British Columbia can be categorized as either commercial facilities that support seawater net-pen aquaculture, or those that supplement naturally occurring Pacific salmon through the Department of Fisheries and Oceans Canada (DFO) Salmonid Enhancement Program (SEP). SEP hatcheries can be subcategorized into federally operated hatcheries and community-operated hatcheries, the latter either in association with the Community Economic Development Program (CEDP) or Public Involvement Program (PIP). For this study, samples were collected from facilities of each category/subcategory distributed across the geographic breadth of British Columbia (
Fig. 1). Kidney and/or spleen tissues were targeted in all sampling events owing to the affinity for PRV to accumulate in these erythrocyte-rich organs (
Garver et al. 2016).
Sampling of commercial hatcheries was conducted as part of the Fish Health Auditing and Surveillance Program performed by DFO Aquaculture Management Division. Sixty fish were targeted in each instance to ensure 95% confidence in identifying at least 5% prevalence of PRV at the population level using the USDA APHIS animal sample size calculator (
https://www.aphis.usda.gov) assuming 95% diagnostic test sensitivity based on performance of similar qPCR assays (
Polinski et al. 2021a;
Yang et al. 2022;
Delphino et al. 2023). Fish were sampled from the general population at all locations as neither morbidity nor outward visual appearance/behavior have shown correlation with PRV infection prevalence or load (
Takano et al. 2016;
Wessel et al. 2017;
Polinski et al. 2019,
2022;
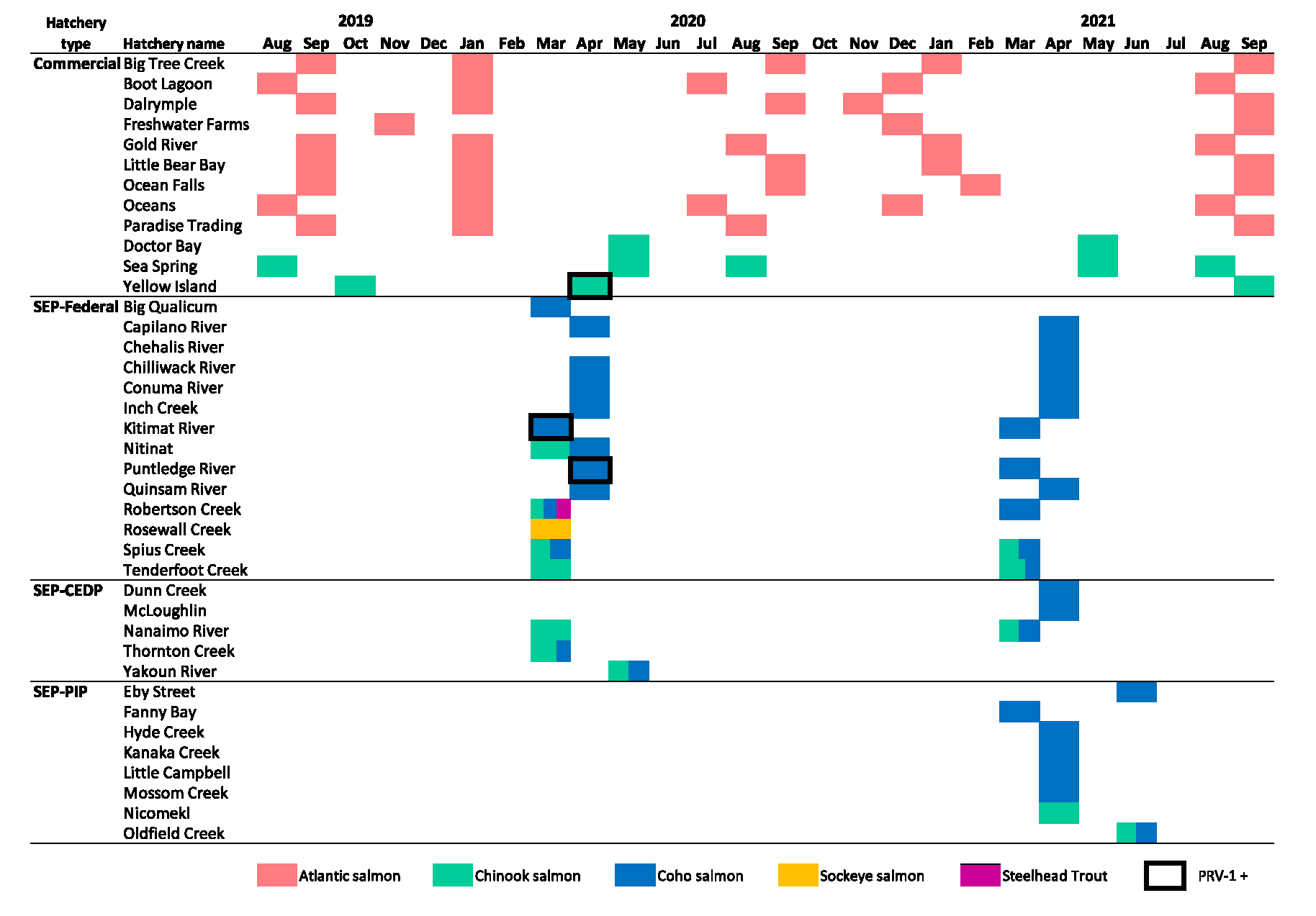
Marty et al. 2020). Samples were collected at all 12 licensed commercial freshwater salmon facilities in British Columbia: 9 that exclusively rear Atlantic salmon and 3 that exclusively rear Chinook salmon (Supplemental material 1). Each facility was visited and sampled 2–6 times between August 2019 and September 2021 culminating in a total of 53 sampling events (
Fig. 2; Supplemental material 1). In each instance, kidney samples (5–50 mg) were collected from 60 fish per sampling event, placed into individual 2 mL microtubes containing 1 mL of RNAlater
® nucleic acid preservation solution and stored at −20 °C. Frozen samples were transported to the DFO Pacific Biological Station (DFO-PBS) in Nanaimo, British Columbia, for processing.
Of the more than 50 SEP freshwater facilities operating in British Columbia, 27 were opportunistically sampled during 39 sampling events between March 2020 and June 2021 (
Fig. 2). This included 14 Federal, 5 CEDP, and 8 PIP facilities based on availability and program interest. Facilities encompassed the full geographic range of SEP in British Columbia (
Fig. 1; Supplemental material 1) and facilities containing Chinook and coho salmon were specifically targeted because (i) overall prevalence of PRV-1 in the northeastern Pacific has been documented to be highest in these species relative to other Pacific salmonids (
Purcell et al. 2018;
Polinski et al. 2020) and (ii) PRV-2 and PRV-3 have also been documented in coho salmon in freshwater from other countries (
Takano et al. 2016;
Cartagena et al. 2020). Samples were collected as close as possible to the end of a production cycle (typically within 1 month of release) to maximize the likelihood of detecting infections that are known to persist in aquaculture-associated populations for at least 14 months once established (
Garver et al. 2016).
All SEP sampling was conducted either by DFO researchers during hatchery visits, or whole fish were shipped directly to DFO-PBS by hatchery personnel. On-site tissue collections by DFO researchers were performed during 15 sampling events at 14 SEP facilities. Tissues (8–22 mg kidney and/or spleen; N = 60 per hatchery sampling event) were collected from MS-222 euthanized fish, individually weighed in 1.2 mL microtubes, and immediately frozen at −80 °C. During five additional sampling events involving five hatcheries, MS-222 euthanized whole fish were collected on-site by DFO researchers and immediately frozen at −80 °C for transport. In the laboratory, specimens were thawed at 4 °C, and kidney and/or spleen tissues excised, weighed, and processed for nucleic acid extraction as described below. For the remaining 19 sampling events conducted across 17 facilities, MS-222 euthanized fish were collected whole by hatchery personnel into bags based on species and stock of origin. Fish were either immediately shipped on ice (1 event) or frozen at −20 °C (18 events) and then shipped to PBS, whereupon fish were thawed at 4 °C and tissues collected as described above.
Nucleic acid extraction and RT-qPCR screening
Nucleic acid extraction and diagnostic detection of PRV RNA was conducted at either the DFO-PBS (5219 samples), the British Columbia Centre for Aquatic Health Sciences (CAHS; 200 samples), or the Okanagan Nation Alliance Fisheries Research and Diagnostic facility (ONA; 200 samples). At DFO-PBS, nucleic acids were purified from tissues either manually using Trizol
® (Thermo-Fisher Scientific; 448 samples) or via a semiautomated process involving a Kingfisher Flex and MagMAX™ Pathogen RNA/DNA kit (Thermo-Fisher Scientific; 4771 samples). Eluted RNA (up to 1 µg) from either method was then reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and screened for PRV-1 (
Polinski et al. 2022). A detailed protocol for nucleic acid extraction, reverse transcription, and PRV-1 qPCR screening is provided in Supplementary material 2. A portion of extracted RNA from a subset of samples (2205 samples encompassing all five salmon species and all five subcategories of freshwater facilities; Supplementary material 1) was additionally screened for PRV-1, -2, and -3 using a Pan-PRV assay as previously described (
Zhao et al. 2021). Absolute PRV quantification was determined in each instance by serial dilution of a 482 bp double-stranded DNA gBLOCK fragment (Integrated DNA Technologies) consisting of sequence targeted by the qPCR primer and probe (
Garver et al. 2016;
Zhao et al. 2021). A six-step 10-fold dilution series of the gBLOCK fragment spanning a dynamic range of 100–10
7 target copies per reaction was incorporated in duplicate into each run.
Sample screening conducted at CAHS was performed using the same methods as presented for DFO-PBS (200 samples via MagMAX™ extraction; Supplementary material 1). At ONA, nucleic acid was purified from tissues using Quick-DNA/RNA Pathogen Mag Bead extraction kit (Zymo Research) according to the manufacturer's recommendations. Eluted RNA was reverse transcribed using Quantitech™ reverse transcription kit (Qiagen) and screened for PRV1 qPCR using QuantiNova Probe PCR kit (Qiagen). A detailed protocol for nucleic acid extraction, reverse transcription, and PRV-1 qPCR screening is provided in Supplementary material 2.
Reproducibility for PRV-1 detection
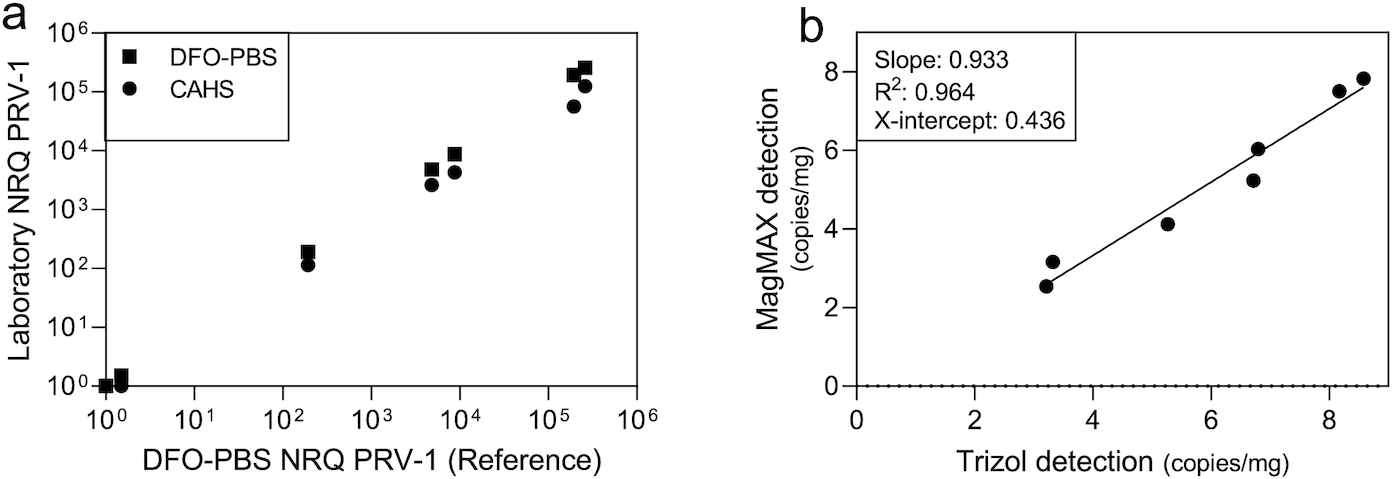
Consistency in PRV-1 detection between laboratories and processing methods was determined by comparing results of replicate kidney samples from 10 farmed Atlantic salmon with predetermined PRV status (7 positive, 3 negative) tested at both DFO and CAHS laboratories. A replicate kidney sample was sent to both laboratories and all sample processing was completed blinded to prescreening PRV status. Additionally, the 10 samples sent to DFO-PBS were divided prior to nucleic acid extraction and processed separately using both the Trizol® or MagMAX™ nucleic acid extraction protocols. To further confirm interlaboratory agreement, population sets (n = 60 per set) from each of 10 commercial hatchery sampling events were equally distributed and screened at DFO, CAHS, or ONA diagnostic laboratories (i.e., n = 20 samples per laboratory per population set; 600 total samples; Supplementary material 1).
S1 genome sequencing and phylogenetic comparisons
S1 genome sequencing was attempted in the four instances in this study where PRV genomic material was detected at a mean concentration greater than 1000 copies/mg (i.e., Ct ∼ <32), which in our experience represents a minimum concentration necessary for reliable sequence coverage using our methods (data not shown). A portion (1 µg) of purified total RNA not used for qPCR analysis was preamplified using primer pairs designed in Primal Scheme (
http://primal.zibraproject.org) to target the S1 sequence of PRV-1 (Supplementary material 1) and run in multiplex qPCR, cleaned and sequenced following the protocol outlined by
Quick et al. (2017) using a nanopore MinION sequencer. Reads were assembled against the published PRV-1 genome (GenBank GCA_002829625.1;
Kibenge et al. 2013) using NanoPipe (
Shabardina et al. 2019). Phylogenetically comparisons of S1 segments were performed against a select subset of 230 published S1 sequences previously evaluated for phylogeny (
Siah et al. 2020; Supplementary material 1) via a Tamura–Nei neighbor-joining method following Clustal Omega maximum-likelihood alignment using Geneious Prime 2022.1.1.
Discussion
Transmission of PRV (all genotypes) has been readily accomplished in freshwater and has been well documented in freshwater facilities raising juvenile anadromous salmon from North America, Europe, and Japan (reviewed by
Polinski et al. 2020). This has included multiple commercial freshwater Atlantic salmon facilities in British Columbia within the past decade (
Marty et al. 2015;
Garver et al. 2016;
Polinski et al. 2019). Following initial detection of PRV-1 in 2013, commercial Atlantic salmon growers in British Columbia have made concerted efforts toward removing PRV-1 from freshwater facilities. A decreasing trend for PRV-1 prevalence was observed from 2013 to 2018 where 42% of 48 fish sampled from two hatcheries were PRV-1 positive in 2013 and by 2018, 0.2% of 586 fish sampled from eight hatcheries were positive for PRV-1 (
Mimeault et al. 2019). Our findings indicate that this decreasing prevalence trend has continued, and that by 2019 all Atlantic salmon facilities have likely been PRV-1 free. It therefore does not appear that freshwater Atlantic salmon facilities used to seed regional seawater net-pens have played a meaningful role in PRV-1 persistence in the region in recent years even though they may have acted as a reservoir historically within the region. Further, given that some Atlantic salmon producers rely on PRV-1 infected broodstock for gametes (
Polinski et al. 2022), it also appears that current iodine-based disinfection strategies employed by the industry are effective at deterring egg-associated transmission of PRV-1 as previously suggested (
Polinski et al. 2020).
In considering the three commercial Chinook salmon freshwater facilities in British Columbia, this study identified only one individual in one cohort at one facility to be putatively infected with PRV-1. If this detection represents a true positive, it is interesting that this did not appear to create a persistent viral presence in the facility. Laboratory challenge studies with PRV-1 have identified that the virus quickly spreads through a confined population once established, which results in readily detectable persistent infections (
Garver et al. 2016;
Wessel et al. 2017). Yet, repeat sampling of this facility with a subsequent cohort in 2021 did not detect virus. It is therefore possible that the 2020 detection represents a false positive result. Although qPCR has demonstrated both high diagnostic specificity and sensitivity—often greater than 98% specificity in stringently controlled laboratories (
Polinski et al. 2021a;
Yang et al. 2022;
Delphino et al. 2023)—false positive detections become increasingly likely as sample size increases. Further, false positive detections are also most likely to manifest at or near the lower limit of detection of the qPCR assay due to a combination of increased probability for low-quantity aerosol contamination to occur or nonspecific fluorescence changes to manifest in late qPCR cycles (
Polinski et al. 2021a;
Meyers and Hickey 2022). In this study, all three inconclusive detections (1 coho, 1 Chinook, and 1 Atlantic) as well as one positive detection from a Chinook salmon were at or below the lower effective limit of detection of the qPCR assay (i.e., less than 1–3 theoretical copies per reaction; Ct > 35) and represent <0.08% of screened samples, implying that theoretical diagnostic specificity was 99.92% or greater in this study even if these detections were false. It therefore becomes highly speculative in defining inconclusive samples from this study as either infected or not infected with PRV-1, and we therefore make no recommendation for dichotomizing inconclusive results presented here. We further treat the one commercial chinook salmon detection in this instance with substantial uncertainty. Irrespective of the validity of the single chinook detection however, it appears that commercial freshwater Chinook salmon facilities have not been a major contributor to the regional prevalence of PRV-1 within the past few years.
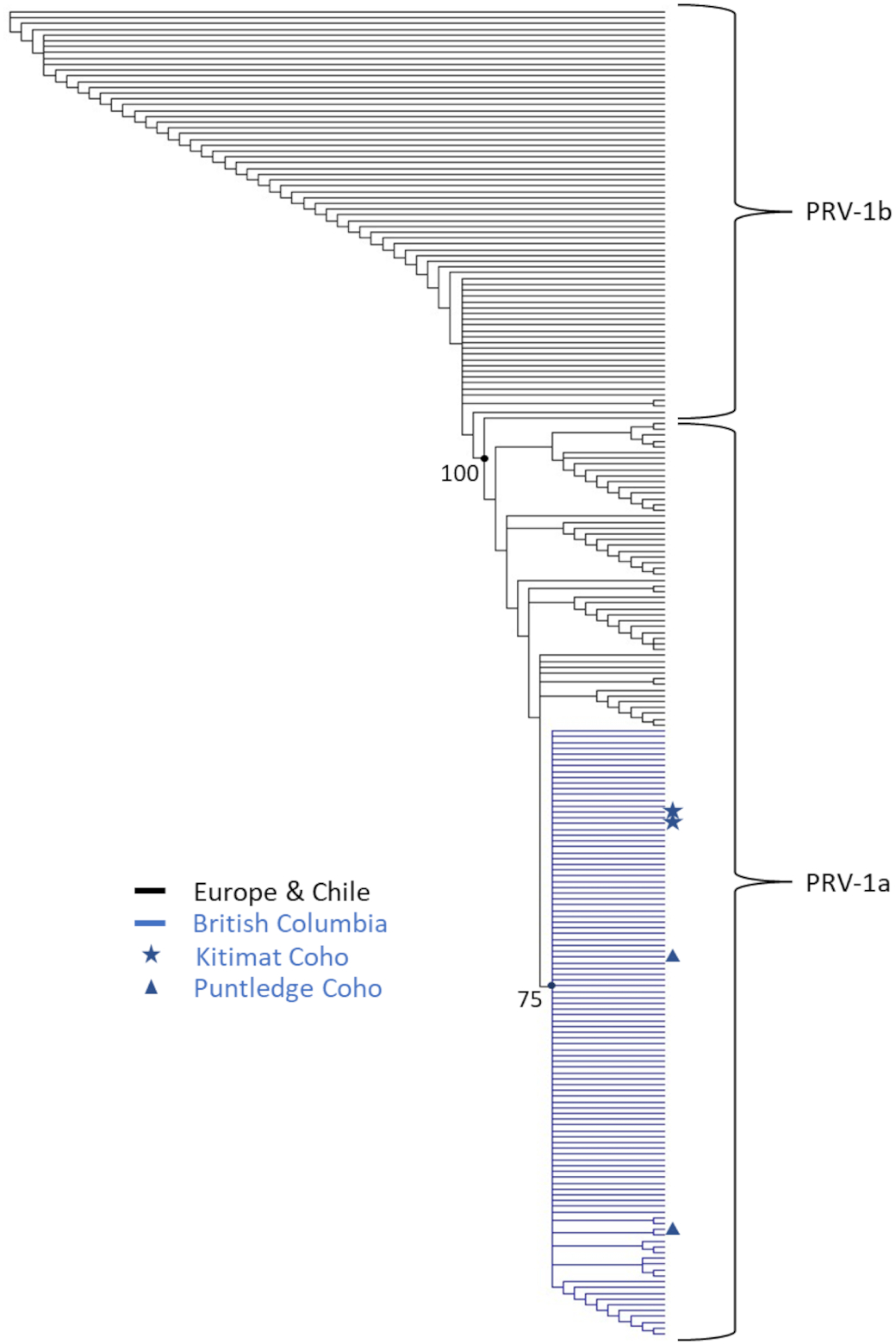
The four coho salmon with PRV-1 infections from SEP facilities in this study all had relatively high viral RNA copy numbers. This, in concert with identifying repeat positive samples at the population level in both premises where PRV-1 was detected and ability to sequence a second viral genomic segment, provides strong evidence for diagnostic accuracy in these instances. Interestingly, the relatively low prevalence for PRV-1 in each of the two sample sets (both 3.3%) suggests that viral transmission within each facility was either very low or the populations had only been recently infected. Cumulative regional prevalence of PRV-1 in marine wild coho salmon has been around 6% (
Polinski et al. 2020) and PRV-1 has been occasionally detected in returning adults to both facilities (S. Johnson (personal communication)), which provides at least one putative infection source. It was also anecdotally noted that detections in 2020 at Kitimat hatchery occurred shortly after fish were transitioned from well water to river water prior to release. The fact that the S1 sequences from the two positive detections at Puntledge hatchery in 2020 were not identical indicates that perhaps multiple infection sources were present at that facility. Whatever the potential source, it appeared to be transitory, as there was no carrying over of infection between 2020 and 2021 year-classes at either facility. Additionally, the low PRV-1 prevalence in these two instances and the lack of detection at all other SEP facilities indicate that the SEP program at large does not appear a major reservoir for PRV-1 in the region. Historic contributions of PRV also seem unlikely from these facilities given that PRV has not been cleared from a local region/facility to our knowledge once it has been introduced without targeted mitigation and monitoring, and SEP facilities in British Columbia have neither monitored nor actively mitigated against PRV transmission.
The absence of PRV-2 and PRV-3 detection within SEP coho during this study is also significant. Both genotypes have been associated with freshwater coho populations in all other regions where the genotypes are endemic (reviewed by
Polinski et al. 2020) and its lack of detection here provides strong evidence that neither of these PRV genotypes are present in the region. However, there is a tropism for PRV-3 to infect rainbow trout in Europe and rainbow trout have a potential ability to clear PRV-3 within 2 or 3 months postinfection (
Hauge et al. 2017;
Vendramin et al. 2019)—a unique circumstance for PRV–salmon interactions. A current lack of screening for PRV-3 in rainbow/steelhead trout in the northeastern Pacific (only 20 steelhead from one facility were screened in this study) thus provides a potential unexplored reservoir for PRV-3 occurrence in the region, although the fact that both steelhead and coho are sometimes reared concurrently in SEP hatcheries makes it seem unlikely that a putative unidentified PRV-3 rainbow/steelhead trout reservoir has not spilled over into cohabitating coho.
To efficiently screen the more than 5000 samples collected in this study, we developed and assessed a semiautomated process for purifying PRV nucleic acid (MagMAX kits using the Kingfisher Flex). This method proved highly similar in sensitivity and performance to previously published detection methods using phenol/guanidine-based methods (Trizol). Although not directly compared in this study, it also appears that quantitative estimations by our new method are reasonably similar to column-based purification procedures (e.g., QIAGEN's QIAamp Viral RNA mini kit) (
Wessel et al. 2020). We anticipate that this method will prove highly desirable in future efforts for screening large population sets for PRV. This may be of particular importance if any of the three PRV genotypes or subgenotypes become nationally or internationally regulated in future.
In conclusion, data generated in this study indicate that commercial and enhancement freshwater salmon facilities of British Columbia currently contribute minimally to the regional ubiquitous seawater prevalence and persistence of PRV-1 and confirm that seawater reservoirs are the primary mechanism for continued PRV-1 persistence in the region. Freshwater facilities also appear not to have contracted or participated in the distribution of nonendemic forms of PRV as neither PRV-2, PRV-3, or Atlantic variants of PRV-1 were detected. Cumulatively, this would indicate that continuation of current aquaculture management strategies and practices in both commercial and SEP freshwater facilities is unlikely to affect regional prevalence of this virus.