Wastewater Surveillance for SARS-CoV-2 RNA in Canada
Abstract
Graphical Abstract
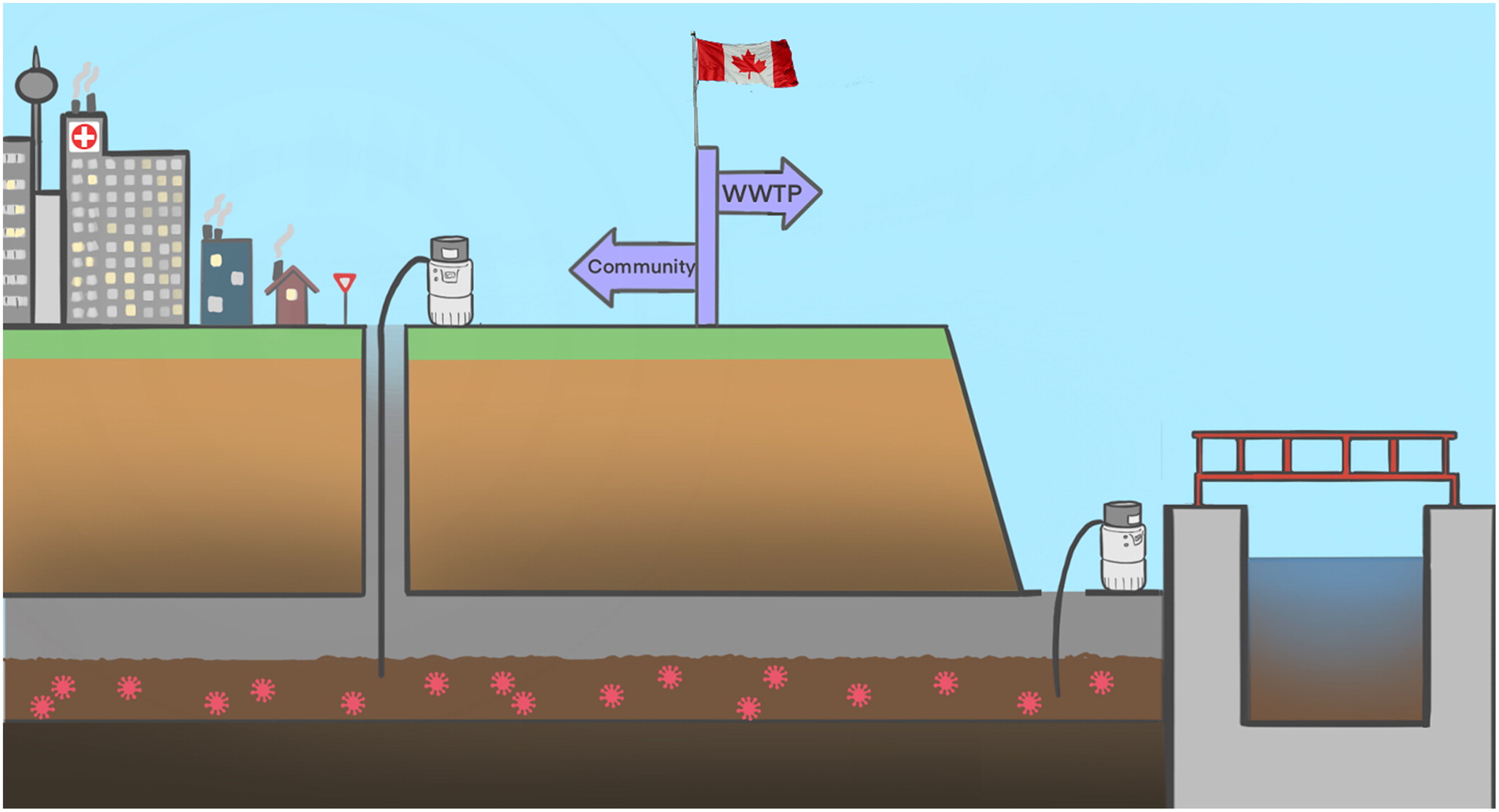
1. Background, purpose, and introduction
1.1. Background
1.2. Purpose for this report
1.3. Pre-COVID applications of wastewater surveillance for public health purposes
1.4. International pursuit of wastewater surveillance for SARS-CoV-2 RNA
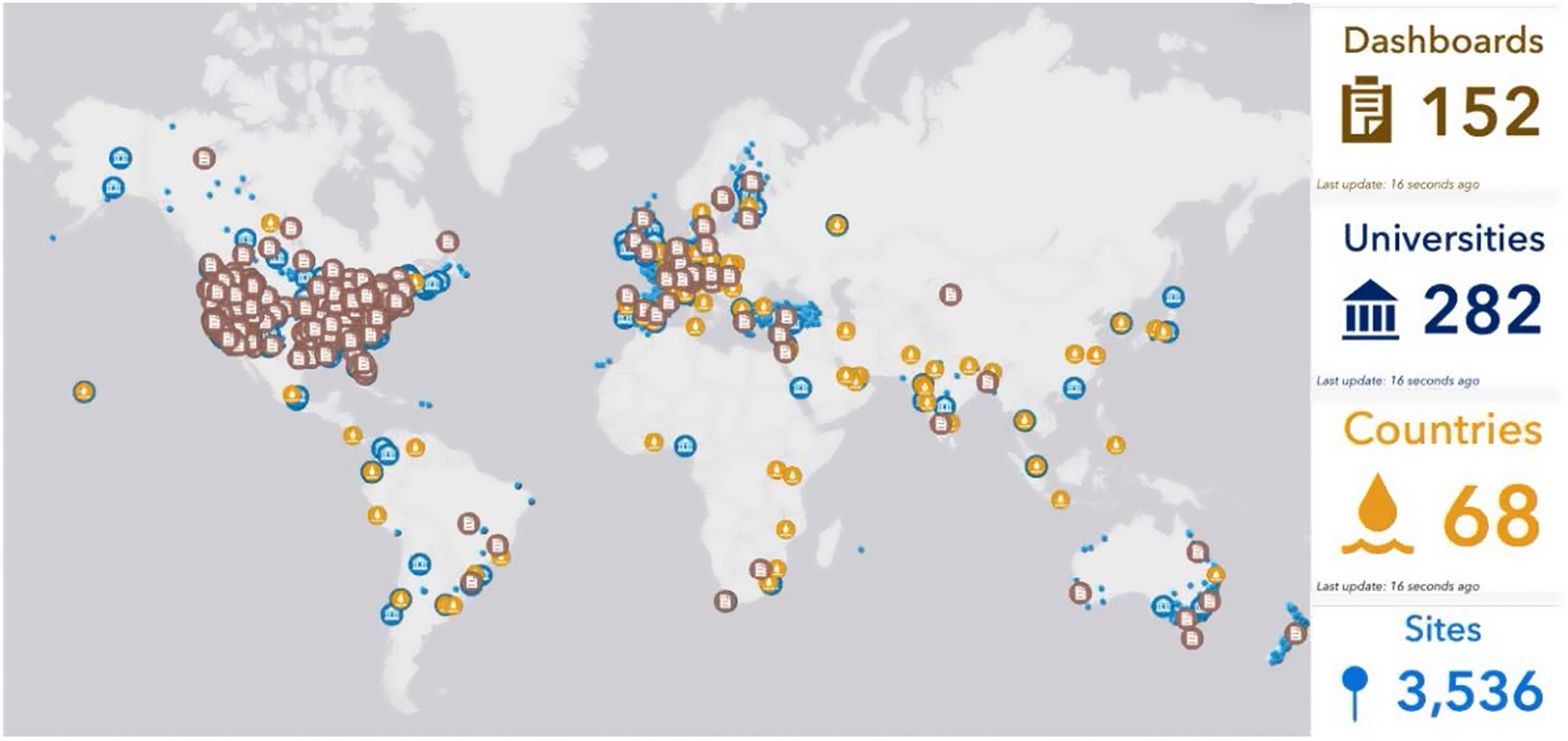
1.5. Initiation of wastewater surveillance for SARS-CoV-2 RNA in Canada
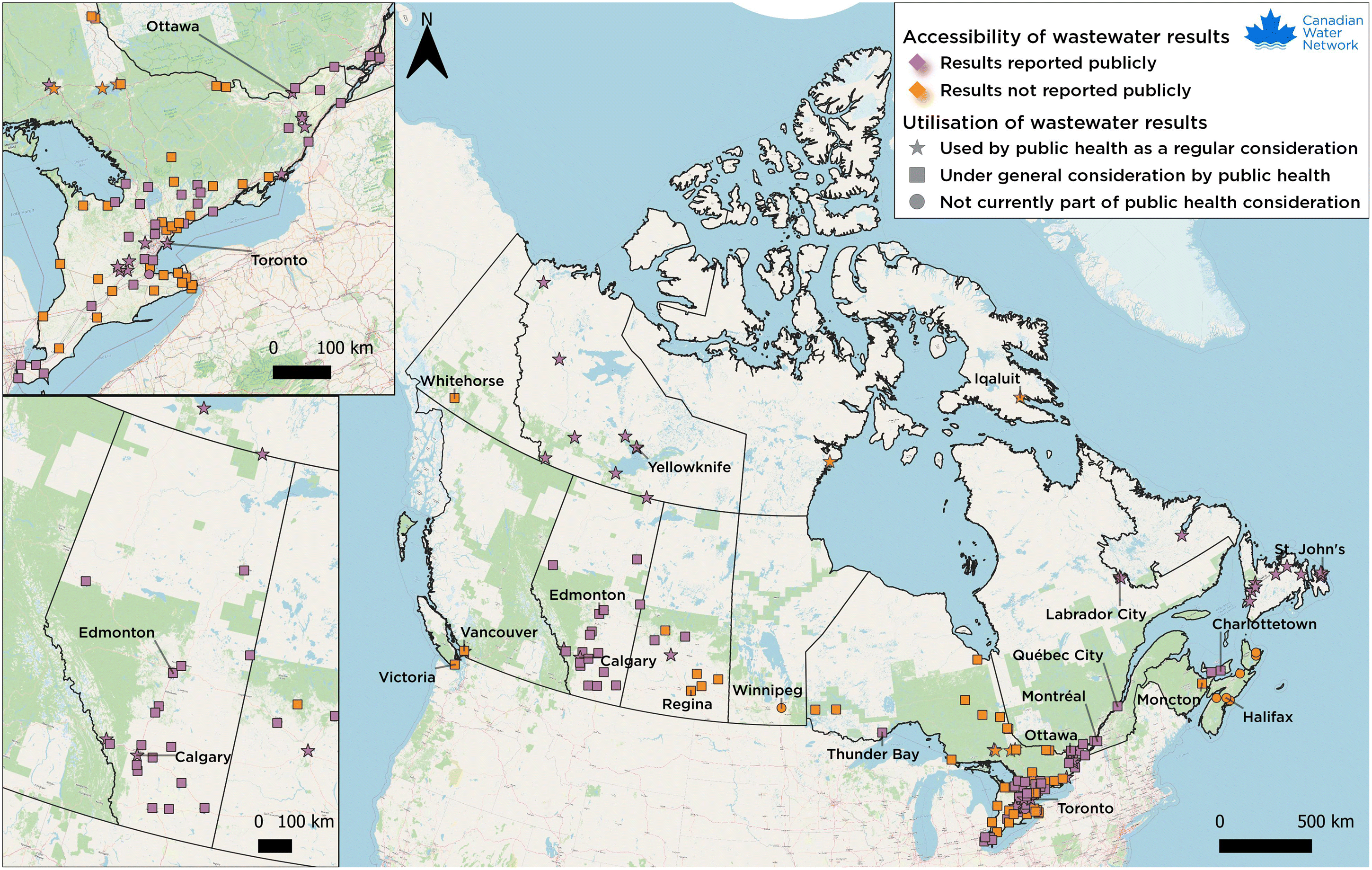
2. Public health measures for a pandemic
2.1. Introduction
2.2. Non-pharmaceutical interventions (NPI) for a pandemic
| Human surveillance & individual preventive measures | Case reporting Early rapid viral diagnosis Disinfection Hand hygiene Respiratory etiquette Surgical and N95 masks Other personal protective equipment |
| Patient management | Isolation of sick individuals Provision of social support services to the isolated |
| Contact management | Quarantine Voluntary sheltering Contact tracing |
| Community restrictions | School closures Workplace closures Cancellation of group events International and domestic travel restrictions |
| Stage of the Pandemic | ||||
|---|---|---|---|---|
| Non-Pharmaceutical Interventions | No cases in Country | Cases in country, none local | Early localized cases | Advanced – wide-spread transmission |
| Hand hygiene - hospital | ||||
| Hand hygiene - ambulatory | ||||
| Hand hygiene - community/home | ||||
| Respiratory etiquette - hospital | ||||
| Respiratory etiquette - ambulatory | ||||
| Human surveillance | ||||
| Case reporting | ||||
| Rapid viral diagnosis and triage | ||||
| Voluntary advisories on departures from international affected regions | ||||
| Voluntary self-isolation of the sick in home | – | |||
| Provision of social support services (to isolated or quarantined persons) - hospital | – | |||
| Provision of social support services (to isolated or quarantined persons) - ambulatory | – | |||
| Other PPE - hospital | ||||
| N95 Respirators - hospital | ||||
| Respiratory etiquette - community/home | ||||
| Surgical masks - hospital | ||||
| Surgical masks - ambulatory | ||||
| Provision of social support services (to isolated or quarantined persons) - home | – | |||
| N95 Respirators - ambulatory | ||||
Note: Legend: Majority recommendation for use ![]() ; Majority recommendation against use
; Majority recommendation against use ![]() ; Disagreement - no majority
; Disagreement - no majority ![]() ; -- not relevant.
; -- not relevant.
| Stage of the Pandemic | ||||
|---|---|---|---|---|
| Non-Pharmaceutical Interventions | No cases in Country | Cases in country, none local | Early localized cases | Advanced – wide-spread transmission |
| N95 Respirators - ambulatory | ||||
| Limited case-by-case home-based mandatory quarantine (of exposed - home) | – | |||
| Contact tracing | ||||
| Mandatory restrictions on arrivals from affected international regions | ||||
| Exit screening of travelers from affected international regions to unaffected U.S. regions | ||||
| Entry screening of travelers from affected international regions to unaffected U.S. regions | ||||
| Exit screening of travelers from affected U.S. regions to unaffected U.S. regions | – | |||
| School closures | – | |||
| Work closures | – | |||
| Case-by-case cancellation of public events | – | |||
| Mandatory restrictions on arrivals from affected U.S. regions | – | |||
| Entry screening of travelers from affected U.S. regions to unaffected U.S. regions | – | |||
| Mandatory restrictions on departures from affected international regions | ||||
| Surgical masks - community | ||||
| Surgical masks - home | ||||
| N95 Respirators - community | ||||
| N95 Respirators - home | ||||
| Mandatory restrictions on departures from affected U.S. regions | – | |||
| Other PPE - community | ||||
| Other PPE - home | ||||
Note: Legend: Majority recommendation for use ![]() ; Majority recommendation against use
; Majority recommendation against use ![]() ; Disagreement - no majority
; Disagreement - no majority ![]() ; -- not relevant.
; -- not relevant.
2.3. Surveillance measures for a pandemic
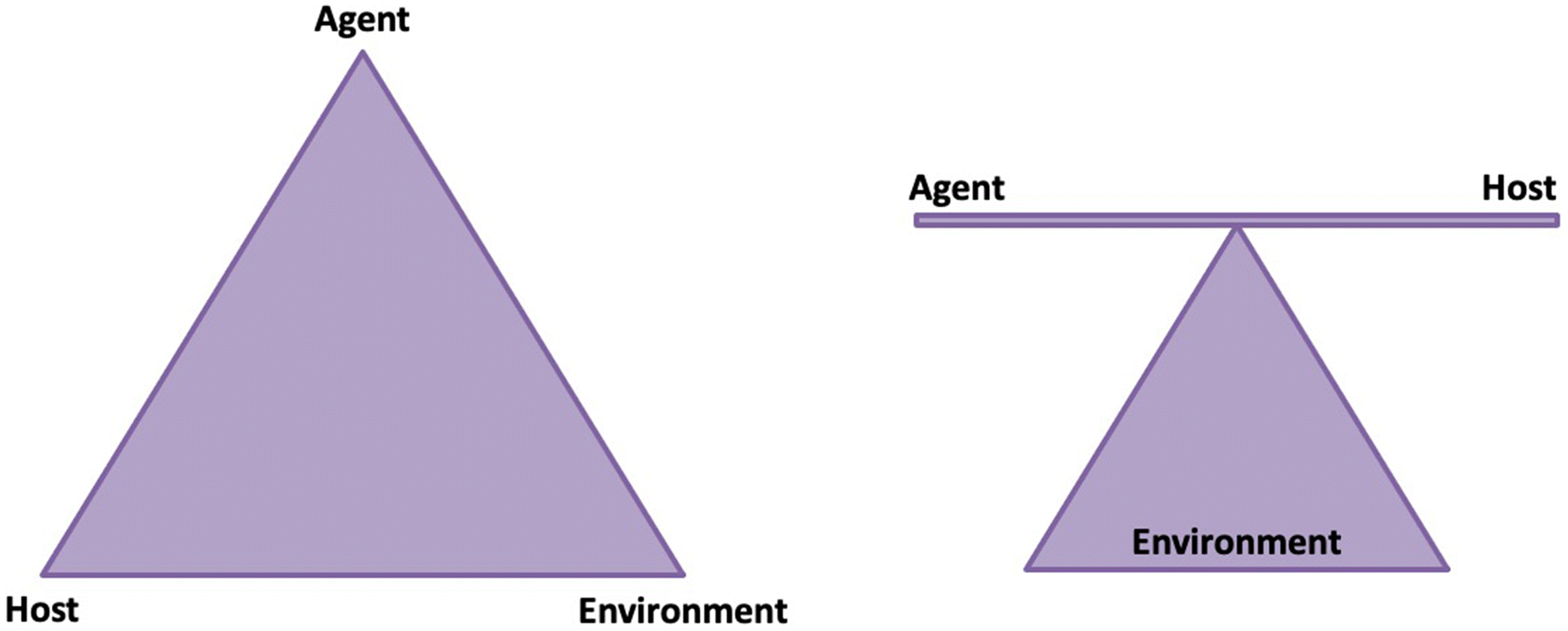
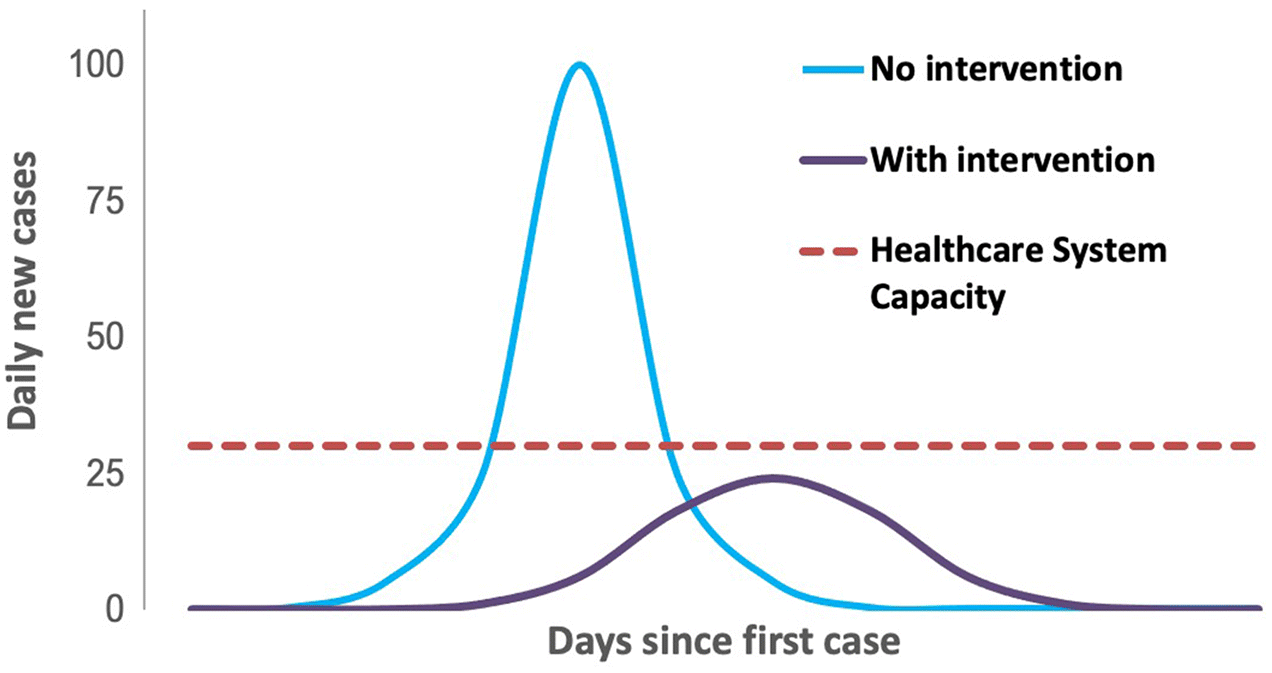
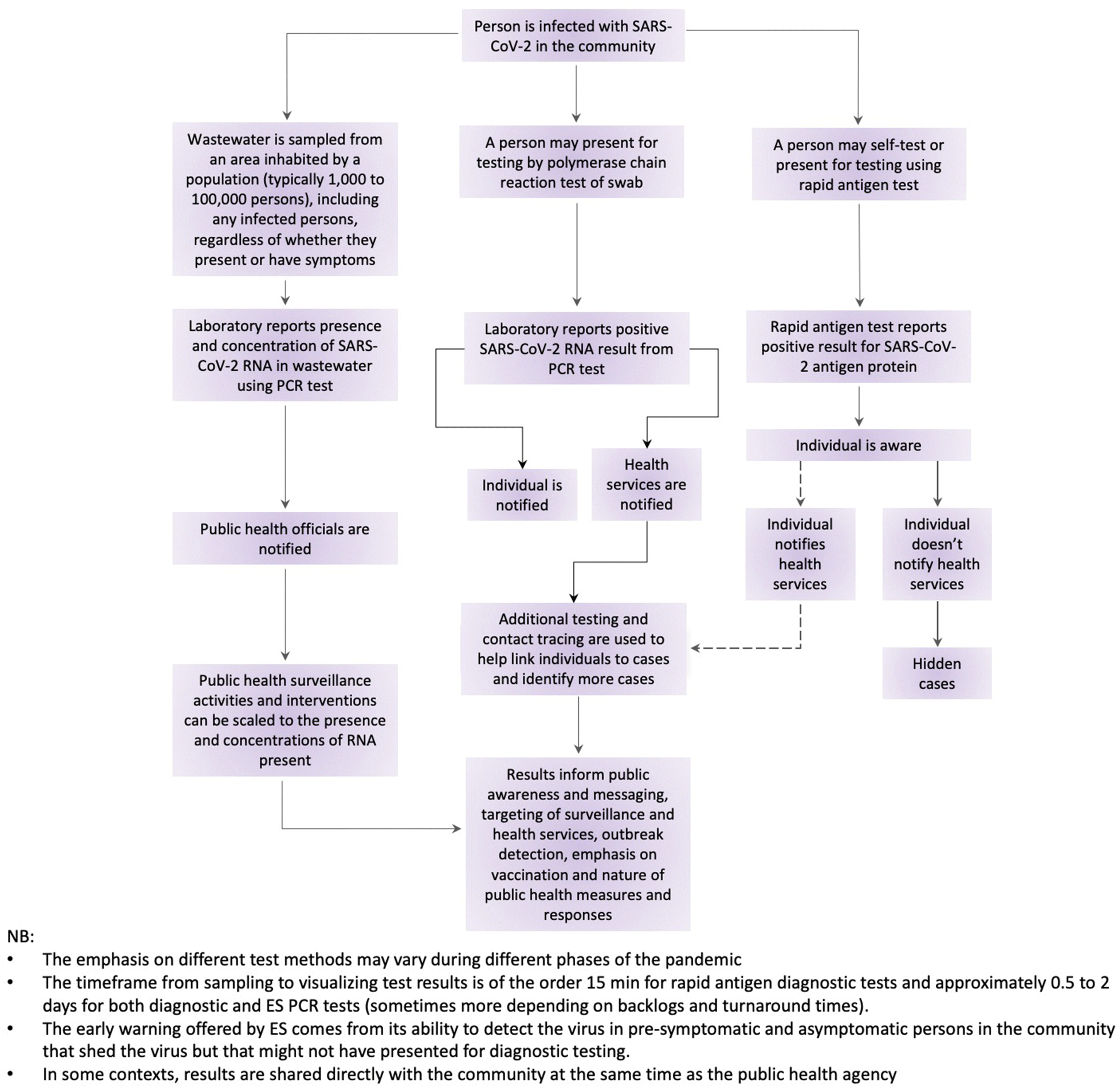
3. Applications of wastewater surveillance for SARS-CoV-2 RNA
3.1. Introduction
3.2. Surveillance at wastewater treatment plants (WWTPs)
3.3. Surveillance in sewer networks to identify COVID-19 cases in neighbourhoods
3.4. Wastewater surveillance of COVID-19 in institutions
3.4.1. Educational institutions
3.4.2. Long-term care facilities and hospitals
3.4.3. Industrial plants and correctional facilities
3.4.4. Surveillance of wastewater to identify COVID-19 cases associated with transportation
3.4.5. Summary
4. Elements of a wastewater monitoring / detection system for SARS-CoV-2
4.1. Introduction
4.2. Wastewater sample collection
4.2.1. Wastewater treatment plants (WWTPs)
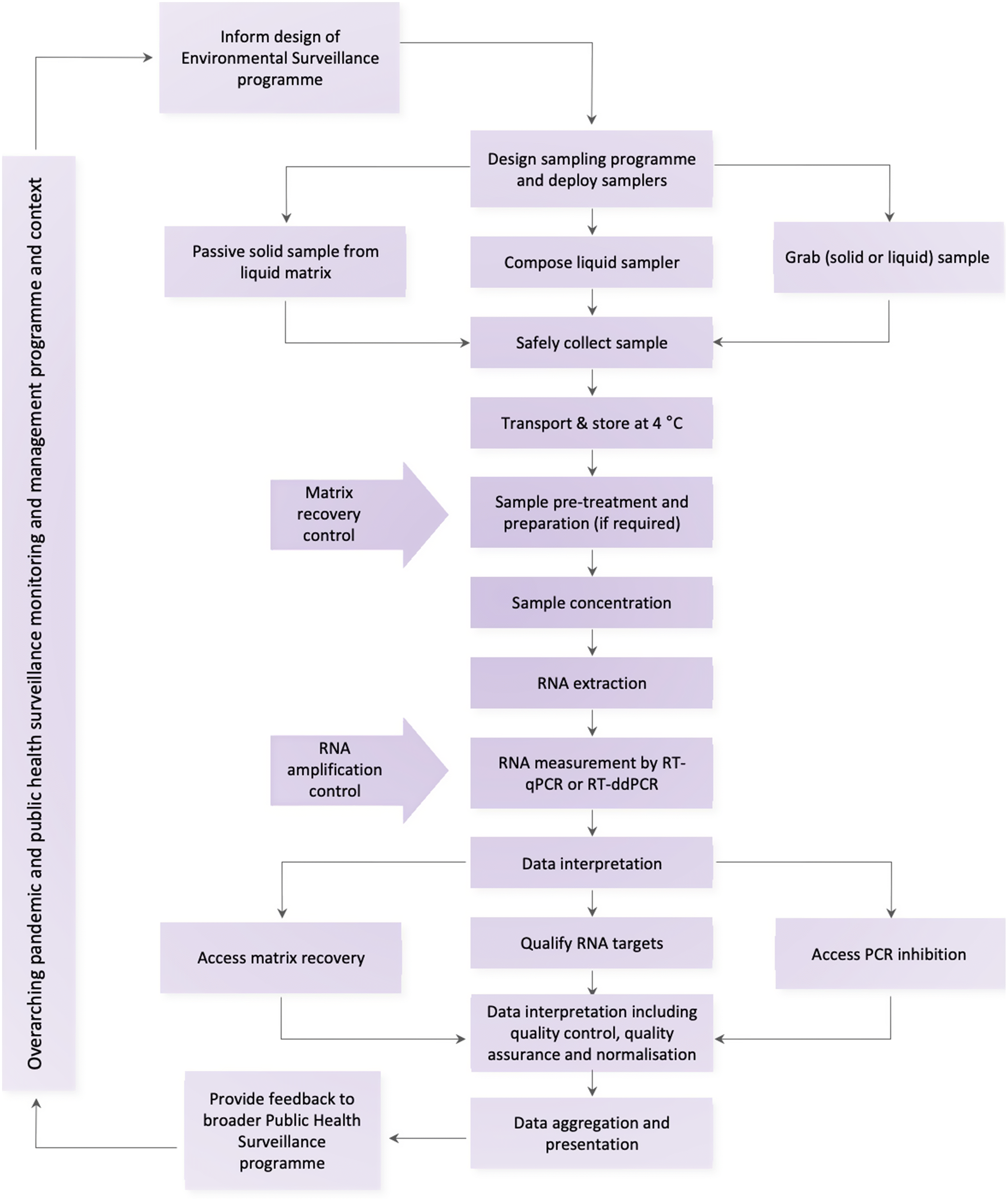
4.2.2. Sewer network sampling
4.3. Wastewater sample preparation and analysis
4.3.1. RNA extraction
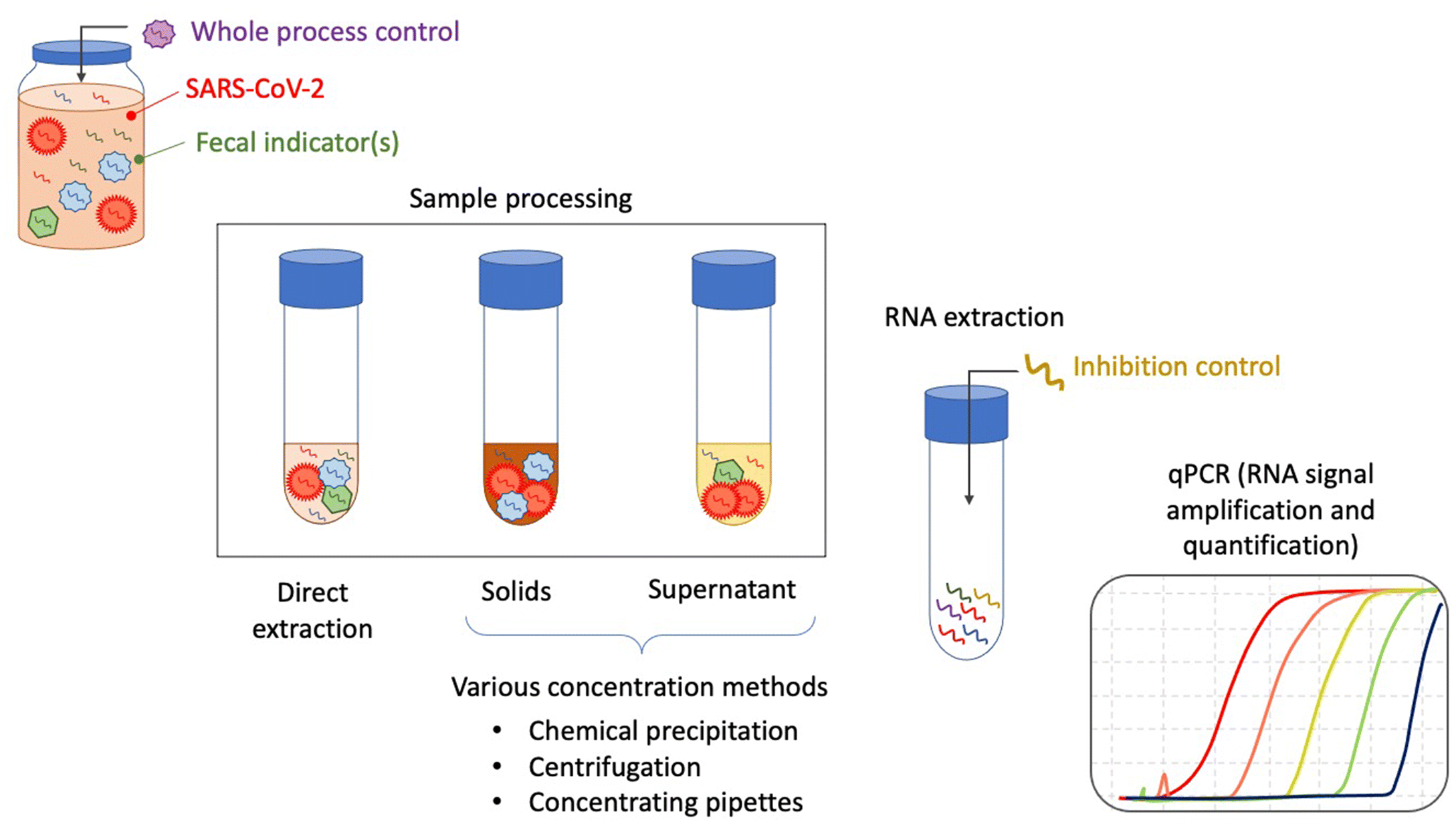
4.3.2. PCR-based detection and quantification
4.3.3. Learning to live with variability
4.3.4. QA/QC verification and validation
4.4. Interpretation of analytical results
4.4.1. Following trends of SARS-CoV-2 RNA levels in community wastewater
4.4.2. Interpreting and understanding SARS-CoV-2 RNA results from wastewater
4.4.3. Normalization of quantitative data to deal with dilution
4.4.4. Communication of results
4.5. Identification and tracking of variants of concern (VOC) in wastewater
4.5.1. How can VOC tracking reduce societal risk and impact of a pandemic?
4.5.2. Which tools are employed to track VOC in Canada?
| WHO Label | Pango Lineage Rambaut et al. (2020a) | Earliest Documentation | Currently circulating in Canada |
|---|---|---|---|
| Variants of Concern | |||
| Alpha | B.1.1.7 | United Kingdom, September 2020 | no |
| Beta | B.1351 | South Africa, May 2020 | no |
| Gamma | P.1 | Brazil, November 2020 | no |
| Delta | B.1.617.2 | India, October 2020 | yes |
| OmicronCurrently includes BA.1, BA.2, BA.3, BA.4, BA.5 BA2.12.1, BA.2.9, BA.2.11, BA.2.13, BA.2.75 sub-lineages and descendent lineages | B.1.1.529 | Multiple countries, November 2021 | yes |
| Variants of Interest | |||
| Epsilon | B.1.427 | USA, March 2020 | no |
| Zeta | P.2 | Brazil, April 2020 | no |
| Eta | B.1.525 | Multiple countries, December 2020 | no |
| Theta | P.3 | Philippines, January 2021 | no |
| Iota | B.1.526 | USA, November 2020 | no |
| Kappa | B.1.617.1 | India, October 2020 | no |
| Lambda | C.37 | Peru, December 2020 | no |
| Mu | B.1.621 | Columbia, January 2021 | no |
4.5.3. Wastewater-based VOC tracking
4.5.4. Challenges and limitations
4.5.5. Today: enhancing situational awareness by strategically employing complementary VOC assays
5. Public health applications of wastewater surveillance and communication needs
5.1. Introduction – challenges and opportunities
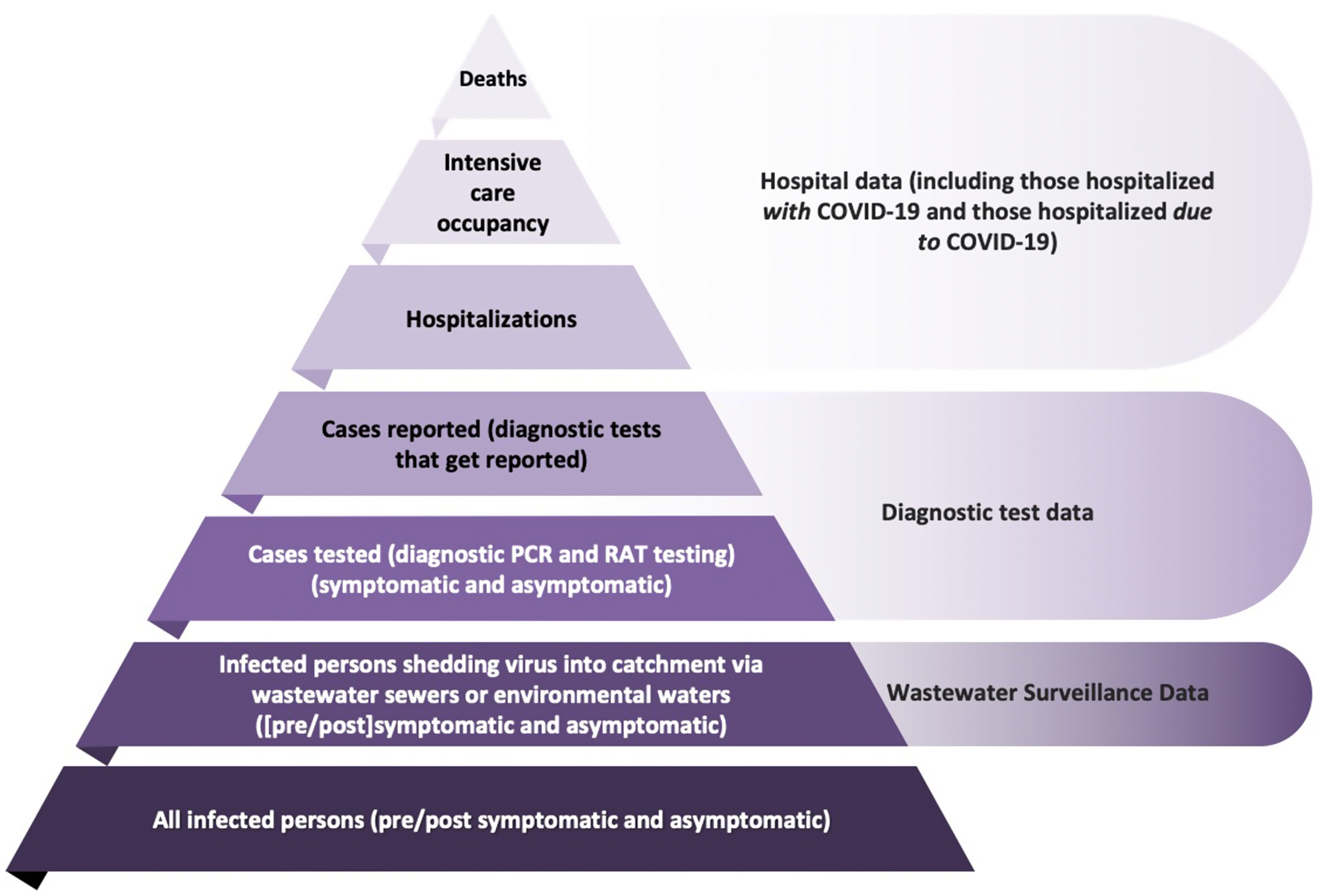
5.2. Detectability of COVID-19 cases by surveillance of SARS-CoV-2 RNA in wastewater
5.3. Early warning and protecting high risk populations
5.4. Tracking trends and concordance of wastewater data with waves of COVID-19 cases
5.5. Tracking of variants of concern (VOCs)
5.6. Modelling to estimate epidemiological indicators
5.7. Ethical considerations
5.8. Public health decision-making
| Benefits for COVID-19 response strategy (Legend +++ = primary benefit, ++ = secondary benefit, + = ancillary benefit) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Application of Wastewater Water Surveillance (use cases) | Description | Provides early warning | Encourages diagnostic clinical testing | Informs decisions on control interventions | Encourages compliance with control interventions | Informs decisions on hospital care capacity | Informs decisions on targeted clinical testing | Improves vaccine uptake | Setting or level where surveillance application has greatest benefit, with comments on benefits |
| Tracking increasing and decreasing Trends at community level to help target COVID-19 responses and interventions | Observing increasing and decreasing trends at community level to, once confirmed, provide an early indication (4-7 days) of changes in incidence & levels of virus circulation for timely decisions on strategies and interventions | ++ | + | +++ | +++ | +++ | Regional & local/city level planning. Applies to all prevalence levels. Communities with low uptake of clinical testing, failing reporting or increased reliance on self-testing. Larger populations sizes | ||
| Finding outbreaks in places thought to be COVID-19 free | Involves testing for SARS-CoV-2 in areas where it is not expected, to provide early warning of its emergence and enable earlier intervention | +++ | +++ | ++ | + | + | Locations where COVID-19 is thought to have been eliminated or locations where COVID-19 cases have not been identified | ||
| Augmenting risk communication to help promote safer behaviours | Publicizing data on detection in wastewater reminds community that the virus is still circulating, may encourage people to seek clinical testing and may reduce complacency about control interventions | + | +++ | + | ++ | + + | Low to moderate prevalence | ||
| Cost-effective targeting of public health surveillance (clinical test resources) | Allows deployment of limited clinical testing resources in hot spot areas with higher signals | + | ++ | ++ | +++ | Spatially differentiated, low to moderate prevalence. Larger population sizes | |||
| Informing early and localized restrictions in pockets of (re-) emergence by helping detect outbreaks | Informs more targeted rapid interventions to minimize the extent and economic impact of restrictions (e.g., service closures, travel limits) | + | +++ | +++ | Spatially differentiated, low prevalence. | ||||
| Identifying existing known Variants of Interest or Concern | Involves testing for known gene targets where proportions of VOCs in circulation are uncertain or higher resolution of information is needed | ++ | ++ | ++ | + | Locations where occurrence of VOCs have not been adequately characterized | |||
| Detecting emergence of VOCs | Involves specialized PCR targets for VOCs or whole-genome sequencing to identify VOCs emerging in the sampled system | +++ | Needs development of specific probes for VOC | ||||||
| Biobanking and Retrospective analysis | Involves retrospective analysis of data to provide intelligence on introduction, evolution & dissemination of the virus to inform future pandemics | ++ | Global, but particularly for areas more vulnerable to future pandemics | ||||||
| Targeted surveillance for early warning of circulation: | Allows early warning to inform earlier intervention to help limit COVID-19 spread in targeted settings | +++ | +++ | ++ | + | ||||
| -vulnerable or high-risk settings | -managed isolation facilities, aged care facilities, schools, prisons, informal settlements, refugees & displaced persons | +++ | +++ | ++ | + | Ensure equity & protect vulnerable groups | |||
| -isolated communities | -remote & indigenous communities, industrial, mining & research facilities; quarantine facilities; student residences | +++ | +++ | ++ | + | Enable “bubbles” or groups to be contained. Augment data in areas with low uptake of diagnostic, clinical testing | |||
| -transport vessels | -sewage tanks of arriving ships & aircraft | +++ | +++ | ++ | + | Test before passengers disembark or disperse | |||
| -multi-day events or gatherings | -meetings, events, or festivals spanning days or weeks | +++ | +++ | ++ | + | Evidence to inform continuation of events or gatherings | |||
Note: Ratings (+, ++, +++) are Interim guidance from WHO, based on experience to early 2022.
5.9. Communications and relations among participants
| Effective Actions | Challenges |
|---|---|
| Establishing early partnerships between the academic research laboratories, municipal utilities, and public health units was critical. e.g., CWN role | Mechanisms to initiate partnerships, especially early in the pandemic. |
| Regular and sustained interaction, reporting and dialogue (interpersonal relationships and trust) | Limitation on human resources to sustain the interactions. |
| Early initiative by academics to test concept and provide early proof of concept. | University laboratory access/ COVID restrictions, lack of personnel, infrastructure and resources. |
| Engaging the public health laboratories at the national and provincial level (e.g., NML, Alberta Provincial Laboratory, BC CDC). | Securing and sustaining commitment and resources early on when agencies were challenged in dealing with the emerging pandemic. |
| Establishing national method leadership at NML (working groups, leadership in method development and application) | Establishing the structure and commitment of Federal Department and Agencies and many provincial governments. |
| Facilitation of regular communication among laboratories and public health agencies nationally to share developments (e.g., informal “coffee club”, PHAC and MECP working groups, CanCOVID) | Need for champions to lead and sustain these initiatives. |
| Open collaboration and sharing of information across laboratories. | Avoiding traditional academic competition and priority concerns for publications, commercial IP, etc. |
| Establishing interlaboratory sample exchanges and studies targeted at improving and contrasting methods | Logistically very difficult and required commitment of resources and personnel. Lack of pre-existing consistent minimum requirements for level QA/QC across laboratories |
| Making data publicly available in almost real time | Concerns about confidence in the interpretation and possible misunderstanding or misuse of complex data by the media or the public |
| Open sharing of data in a common format (e.g.: PHES-ODM; github.com/Big-Life-Lab/PHES-ODM) | Concern over data ownership and rights. Establishing appropriate data-level QA/QC across laboratories. Need for cross disciplinary conversations between laboratories, data scientists/engineers, and data users (epidemiologists) Establishing right data products to serve end user needs |
| Eventual provision of sustained funding from governments to enable the laboratories to have the infrastructure, human resources and material to conduct analysis. | Short term funding because of the uncertainty of scope of the pandemic. Retention of qualified personnel. Moving from academic-based initiative to public health or commercial laboratories |
| Flexibility for research within programs to ensure quality and program development | Early focus of funding surveillance and lack of research funding avenues for academia |
| Provincial data infrastructure and sharing capacity | Logistic, expertise and securing resources |
| Having a champion in public health. (e.g., Peter Juni, the Ontario Science Table. | Finding a champion and a forum within public health agencies. |
| Leveraging existing laboratory quality management Infrastructure (accreditation) | Not in best interest for academic laboratories to seek accreditation, no impetus for commercial laboratories to seek accreditation without a viable, sustainable business case |
| Engaging in open science. Sharing primary data and information and foregoing concerns of institutional advancements. | Difficulties of established institutions to openly share primary data and information can stymie overall progress. |
| High level of public support and media interest. Public engagement with research results, real time questions and answers on social media. High trust of researchers engaging in science communication. | Data transparency and ethical oversight are essential for continued public support. |
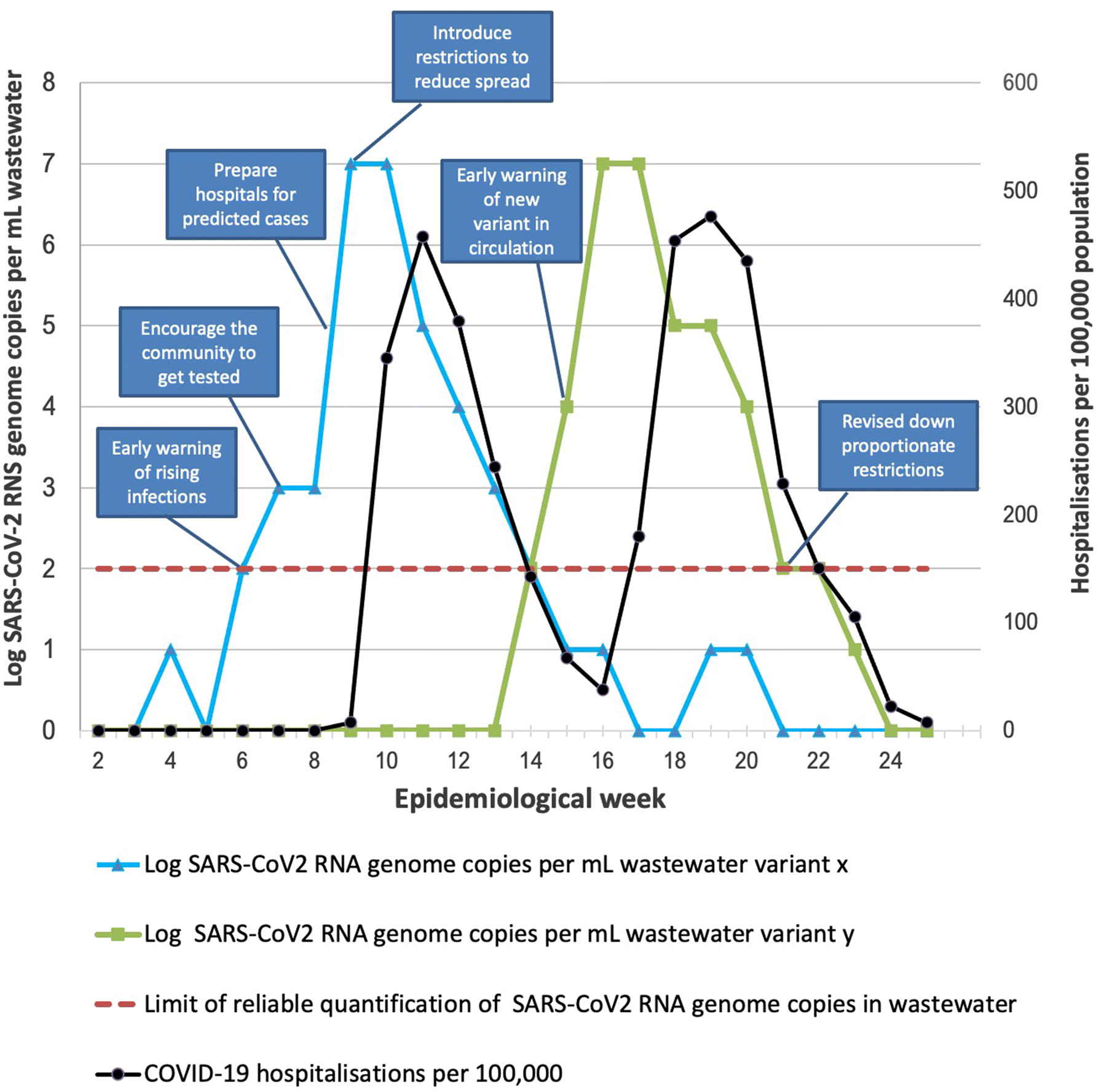
5.10. Some Canadian wastewater surveillance success stories
5.10.1. British Columbia
5.10.2. Alberta
5.10.3. Saskatchewan
5.10.4. Ontario
5.10.5. Québec
5.10.6. Nova Scotia
5.10.7. Newfoundland & Labrador
5.10.8. Public Health Agency of Canada (PHAC) and National Microbiology Laboratory (NML)
6. Strengths and limitations of wastewater surveillance for SARS-Cov-2 RNA
6.1. Strengths of wastewater surveillance for SARS-Cov-2 RNA
6.1.1. Provides objective relevant evidence independent of clinical testing policies
6.1.2. Provides inclusive coverage within a sewershed
6.1.3. Capable of detecting signal from asymptomatic and pre-symptomatic cases
6.1.4. Cost effective sampling
6.1.5. Scalable
6.1.6. Provides useful information on trends
6.1.7. Incorporation into high-level public health risk classifications
6.1.8. Can detect local hotspots and monitor institutions
6.1.9. Tracks dynamics of variants of concern (VOCs)
6.1.10. Ability to document spatial and temporal patterns of virus shedding
6.1.11. Ability to deal with rapid increases in cases that overwhelm clinical testing
6.1.12. Raises public awareness
6.1.13. Non-Invasive surveillance sampling
6.1.14. Generates a valuable database for retrospective analyses
6.2. Limitations of wastewater surveillance for SARS-Cov-2 RNA
6.2.1. Requires accurate knowledge of served population relative to clinical testing
6.2.2. Practical limits of ability to estimate prevalence of COVID-19
6.2.3. Achieving early warning depends on surveillance program factors
6.2.4. Ethical issues
6.2.5. Homes that are on septic systems are generally not covered
6.2.6. Variant tracking requires some specialized analytical capability
6.2.7. No common metric for reporting results
6.2.8. Communication gaps exist among relevant disciplines
6.2.9. Practical limitations for sampling sites in sewer networks
6.2.10. Reticence to participate by some municipalities
6.2.11. High levels of variability in quantitative RNA signals within and between sites
6.2.12. Lack of a coherent strategy for sampling location selection
7. Emerging opportunities and research needs
7.1. Expanding public health applications beyond COVID-19
7.2. Improving analytical methodology
7.3. Developing applications for incorporating wastewater surveillance for SARS-CoV-2 RNA into routine public health surveillance
7.4. Retrospective analyses of wastewater surveillance concerning public health outcomes
7.5. Retrospective analyses of site-specific applications
7.6. Improved methods for normalizing wastewater strength
7.7. Quantitative evaluation of SARS-CoV-2 load and dynamics from sputum vs. faeces
7.8. Attempts at estimating disease prevalence from wastewater require
7.9. Review, evaluation, and development of various models proposed for using wastewater surveillance
7.10. Improving communication and interaction between water utilities and public health
7.11. Value for informing the public for personal risk management
7.12. Development and validation of new methods for surveillance of travel
7.13. Applications to vulnerable communities
7.14. Equitable and representative sampling designs
7.15. Impact of community water use
7.16. Creation of a framework for the use of wastewater surveillance results
8. Conclusions and recommendations
8.1. Conclusions
8.2. Recommendations
Competing interests
Acknowledgements
List of Abbreviations and Terminology
- Allele
- An allele is one of two or more versions of a nucleotide sequence (e.g., a single nucleotide variation in the SARS-CoV-2 genome).
- (AS)-(RT)-(q)PCR
- (Allele-specific) (reverse transcriptase) (quantitative) polymerase chain reaction. Allele-specificity can be added to RT-qPCR which is useful for detection and quantification of VOCs and VOIs.
- Biomarker
- A measurable substance (generally a molecule) that is (or has been) present in an organism and that can be used as an indicator of biological processes, disease processes, or pharmacologic responses to a therapy. Certain biomarkers in environmental samples can detect changes in the degree or extent of disease in a population.
- Copies
- RNA representing a single copy of the viral RNA.
- crAssphage
- Cross-assembly bacteriophage. A bacterial virus (bacteriophage) that infects Bacteroides, a bacterial genus found in the human intestinal tract.
- CT or Ct
- The number of amplification cycles using quantitative reverse transcription Polymerase Chain Reaction (qRT-PCR) technology required for the signal associated with a PCR product (i.e., the target/amplicon) to be detected above a baseline signal that would be present in the assay regardless of whether the target is present (CWN 2020b).
- dd/d PCR
- Digital droplet/digital polymerase chain reaction. A newer iteration of qPCR that relies on partitioning of the sample such that large numbers of small PCR reactions are carried out in parallel that allows more precise calculation of absolute copies of starting material. Analogous to RT-qPCR, RT-dd/dPCR denotes the iteration of the assay used to quantify RNA in a sample.
- DNA
- Deoxyribonucleic acid. The molecule that carries genetic information for the development and functioning of an organism. DNA is made of two covalently linked polymers of nucleic acid that wind around each other to resemble a twisted ladder — a shape known as a double helix (CWN 2020b).
- gBlock
- A synthetic DNA double helix used as a reference material in q/dPCR assays.
- Incidence
- The number of new cases of illness commencing, or of persons falling ill, during a given period in a specified population (Porta 2008).
- NPI
- Non-pharmaceutical Interventions refer to measures such as quarantine, physical distancing, mask-wearing, avoidance of crowds, etc, that do not involved reliance of administration of medicines, as elaborated in Table 1, Section 2.2
- PCR Inhibition
- Inhibitory substances may be present that impede or prevent PCR from running efficiently or effectively, ultimately resulting in delayed Ct quantification (higher Ct) for the actual target of the analysis. Inhibition effects can be monitored by comparing the number of cycles required for detecting a target in a spiked sample matrix compared to that of a distilled water control spiked at the same concentration. Alternatively, inhibition can be inferred if non-linearity of calculated copy number is observed with sample dilution (i.e, no PCR inhibition is present if a sample diluted by ½ results in half the calculated copy number) (CWN 2020b).
- Prevalence
- A measure of disease occurrence: the total number of individuals who have an attribute or disease at a particular time (Porta 2008)
- Positivity Rate
- The percentage of diagnostic tests of a given population tested who test positive for the infection or illness under study (e.g., SARS-CoV-2).
- Reproductive Number
- The basic reproductive number (R0) is used to measure the transmission potential of a disease. It is the average number of secondary infections produced by a typical case of an infection in a population where everyone is susceptible. The effective reproductive number (R, Rt or Re) is the average number of secondary cases per infectious case in a population made up of both susceptible (e.g., unvaccinated) and non-susceptible (e.g., vaccinated) hosts. If R>1, the number of cases will increase, such as at the start of an epidemic. Where R=1, the disease is endemic, and where R<1 there will be a decline in the number of cases. healthknowledge.org.uk/public-health-textbook/research-methods/1a-epidemiology/epidemic-theory
- N1, N2
- Refers to the nucleotide sequences that are commonly amplified by PCR in both clinical and wastewater diagnostic testing for the presence of SARS-CoV-2. It can also refer to the US CDC-designed N1 and N2 PCR assays which are the gold standard diagnostic assays that target the sequences of the nucleocapsid gene of the virus.
- N-gene
- A gene that is present in SARS-CoV-2 genomic RNA and which encodes the nucleocapsid protein that surrounds and protects the genomic RNA present in each infectious viral particle.
- Normalization
- Transformation of the raw SARS-CoV-2 RNA PCR signal to account for systematic variability. Usually, by dividing by another measured factor (e.g., faecal indicator such as PMMoV, flow, TSS, etc).
- NPV
- Negative predictive value (NPV) is the conditional probability: Given that if there is no true case of COVID-19 that the wastewater will correctly report no detectable signal for SARS-CoV-2 RNA.
- NRT
- No-RT control. Used to ensure the observed PCR signal is RNA-dependent (generally used to troubleshoot contamination issues).
- NTC
- No-template control. Used to detect RNA or DNA contamination in the PCR reaction.
- Outliers
- Samples that are statistically not consistent with the other data
- PCR
- Polymerase chain reaction - The process through which genetic material (DNA) can be amplified exponentially through multiple cycles of denaturing, annealing and extension that allows the DNA to self-replicate.
- PCR Efficiency
- One of several performance measures and quality controls for qPCR assays. A low efficiency assay risks under- or over-estimating copies of starting material.
- PMMoV
- Pepper Mild Mottle Virus, a plant virus that infects peppers and other vegeables and is found in feces of humans because it is in the human diet
- PPV
- Positive predictive value (PPV) is the conditional probability given that if there is a true case of COVID-19 that the wastewater will correctly report a positive signal for SARS-CoV-2 RNA.
- QA/QC
- Quality assurance/quality control: A component of quality management focused on providing confidence that specific quality requirements will be fulfilled, and the fulfillment of the requirements specified; relates to how a process is performed to ensure quality requirements are met and the subsequent inspection aspect of quality management.
- RNA
- Ribonucleic acid. A biological molecule that has structural similarities to DNA and serves to direct and enable gene expression.
- RT
- Reverse transcriptase – see explanation below for RT-qPCR
- RT-qPCR
- PCR is a technology platform by which genetic material (DNA) can be amplified exponentially through multiple cycles of denaturing, annealing and extension that allows the DNA to self-replicate. qPCR is an iteration that allows for absolute measurement of copies of starting material. “RT-qPCR” refers to an iteration that includes reverse transcriptase and can thereby measure RNA as a starting material. healthknowledge.org.uk/public-health-textbook/research-methods/1a-epidemiology/epidemic-theory
- Sensitivity
- The proportion of truly diseased people in a population who are identified as diseased by a screening test. Also known as a true positive rate. For an analytical procedure, sensitivity is the conditional probability: that when the target analyte is present, the screening test will detect the analyte’s presence.
- Sewershed
- A segment of a sewer network defined that has the sewers draining into it defined sufficiently well that wastewater samples taken from it can be assumed to represent a defined segment of the sewer drainage system
- Specificity
- The proportion of truly non-diseased people in a population who are identified as non-diseased with a screening test. Also known as a true negative rate. For an analytical procedure, specificity is the conditional probability: that when the target analyte is absent, the procedure will report it as non-detectable.
- Surrogate
- A spike of a related virus (i.e., HCoV-229E or MHV) used as a whole process control
- VOC
- Variant of concern: A SARS-CoV-2 variant that meets the definition of a VOI (see below) and, through a comparative assessment, has been demonstrated to be associated with one or more of the following changes at a degree of global public health significance: Increase in transmissibility or detrimental change in COVID-19 epidemiology; OR Increase in virulence or change in clinical disease presentation; OR Decrease in effectiveness of public health and social measures or available diagnostics, vaccines, therapeutics (WHO 2022b)
- VOI
- Variant of interest: A SARS-CoV-2 variant : with genetic changes that are predicted or known to affect virus characteristics such as transmissibility, disease severity, immune escape, diagnostic or therapeutic escape; AND Identified to cause significant community transmission or multiple COVID-19 clusters, in multiple countries with increasing relative prevalence alongside increasing number of cases over time, or other apparent epidemiological impacts to suggest an emerging risk to global public health (WHO 2022b).
- WWTP
- wastewater (sewage) treatment plant
References
Supplementary material
- Download
- 5.00 MB
Information & Authors
Information
Published In

History
Notes
Copyright
Key Words
Sections
Subjects
Plain Language Summary
Authors
Author Contributions
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
There are no citations for this item
