Freshwater fisheries monitoring in northern ecosystems using Indigenous ecological knowledge, genomics, and life history: Insights for community decision-making
Abstract
Introduction
Past research
Hypothesis
Methods
Study area
IEK and WSD complementarity
| Method to address the parameter and temporal and spatial scale for that parameter | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fish | IEK question | Spatial scale | Temporal scale | Study | WSD | Dataset | Spatial scale | Temporal scalea | Study | Initial prediction |
| Parameter—Population structure/ degree of philopatry/gene flow | ||||||||||
| W | Do walleye look the same throughout the lake? Where do walleye spawn? When do walleye spawn? | Lake-wide | Single snapshot, contemporary perspective (2018) | 1 | ADMIXTURE, global ancestry, at spawningb Pairwise FST, gene flow between populations, (Genodive) | GBS, 8457 SNPs GBS, 8658 SNPs | Four key spawning rivers Four key spawning rivers | Single snapshot, cumulative since colonization (2003, 2015) Single snapshot, cumulative since colonization (2003, 2015) | 2 2 | COM |
| LT | Morphology—variety of morphs described Do you know where and when lake trout spawn? If yes, can you describe/show their location? If yes, do all lake trout that you described earlier spawn in the same location? | Lake-wide | Single snapshot, contemporary perspective (2013) | 3 | ADMIXTURE, global ancestry, feeding seasonb Pairwise FST, gene flow between populations, (Genodive) | GBS, 5830 SNPs GBS, 5830 SNPs | Lake-wide, feeding habitat Lake-wide, feeding habitat | Single snapshot, cumulative since colonization (2013) Single snapshot, cumulative since colonization (2013) | 1 1 | COM |
| NP | Do pike look the same throughout the lake? Where do pike spawn? When do pike spawn? | Lake-wide | Single snapshot, contemporary perspective (2018) | 1 | ADMIXTURE, global ancestry, outside of spawning periodb Gene flow between genetic clusters, pairwise FST (Genodive) | GBS, 245 SNPs GBS, 245 SNPs | Lake-wide, feeding habitat Lake-wide, feeding habitat | Single snapshot, represents cumulative since colonization (2018) Single snapshot, represents cumulative since colonization (2018) | 1 1 | COM |
| Parameter—Body size or morphotypes | ||||||||||
| W | Do walleye look the same throughout the lake? How big, on average, are the walleye that you capture now? | Lake-wide, some specific regions given | Single snapshot, contemporary perspective (2018) | 1 | Lsmeans of total length for each population (lsmeans, R) | Total length (± 1 mm), and mass (± 50 g) | Four key spawning rivers | Single snapshot (2002/2003, 2015) | 2 | COM |
| LT | Morphology—variety of morphs discussed for population structure | Lake-wide | Single snapshot, contemporary perspective (2013) | 3 | Morphology | Relative warp scores for head and body shape | Lake-wide, feeding habitat | Single snapshot (2013) | 4 | CON |
| NP | Do pike look the same throughout the lake? How big, on average, are the pike that you capture now? | Lake-wide | Single snapshot, contemporary perspective (2018) | 1 | Total length and mass for each population | Total length ± 1 mm | Lake-wide, feeding habitat | Single snapshot (2018) | 1 | CON |
| Parameter—Conservation status | ||||||||||
| W | Has the number of fish changed? Has the size of the fish changed? Do you have any concerns about the overall health of the fish throughout the lake? Factors that could contribute to change? | Lake-wide | 5–25 years (2018)c | 2 | Change in total length, mass, length at age over time, Ne, (LDNe); Ho (stacks) | Total length (± 1 mm), mass (± 50g); GBS, 8658 SNPs | Four key spawning rivers | For length/mass, time-series; size-at age, single time-span, 2002–2017; for Ne, single snapshot over 1–2.5 generations (2003–2015) | 2 | COM |
| LT | Have the number of lake trout decreased, increased, or remained the same over the years you have been fishing? Have you noticed any changes to lake trout on the lake since you’ve been fishing? If yes, do you know what might be causing these changes? Do you have any overall concerns about the health of lake trout in Mistassini Lake? What factors might contribute to short-term changes to the number of lake trout there are? | Lake-wide | 20–40 years (2013)c | 1, 3 | Ne (LDNe) | GBS, 5830 SNPs | Lake-wide, feeding habitat | Single snapshot over one generation (2013) | 1 | NAd |
| NP | Has the number of fish changed? Has the size of the fish changed? Do you have any concerns about the overall health of the fish throughout the lake? Factors that could contribute to change? | Lake-wide | 5–25 years (2018)c | 1 | Ne (LDNe) | GBS, 245 SNPs | Lake-wide | Single snapshot over one generation (2018) | 1 | NAd |
Note: Source studies: 1. This study. 2. Bowles et al. (2020). 3. Marin et al. (2017). 4. Marin et al. (2016). LT, lake trout; GBS, genotyping-by-sequencing; SNP, single nucleotide polymorphism; COM, complementarity of results between IEK and WSD; W, walleye; NP, northern pike; CON, congruence of results between IEK and WSD; Ne, effective population size; LDNe, software used; Ho, observed heterozygosity.
Indigenous ecological knowledge
Western scientific data
Fish sampling and sample selection
Sequencing, bioinformatics and statistics
Results
Walleye
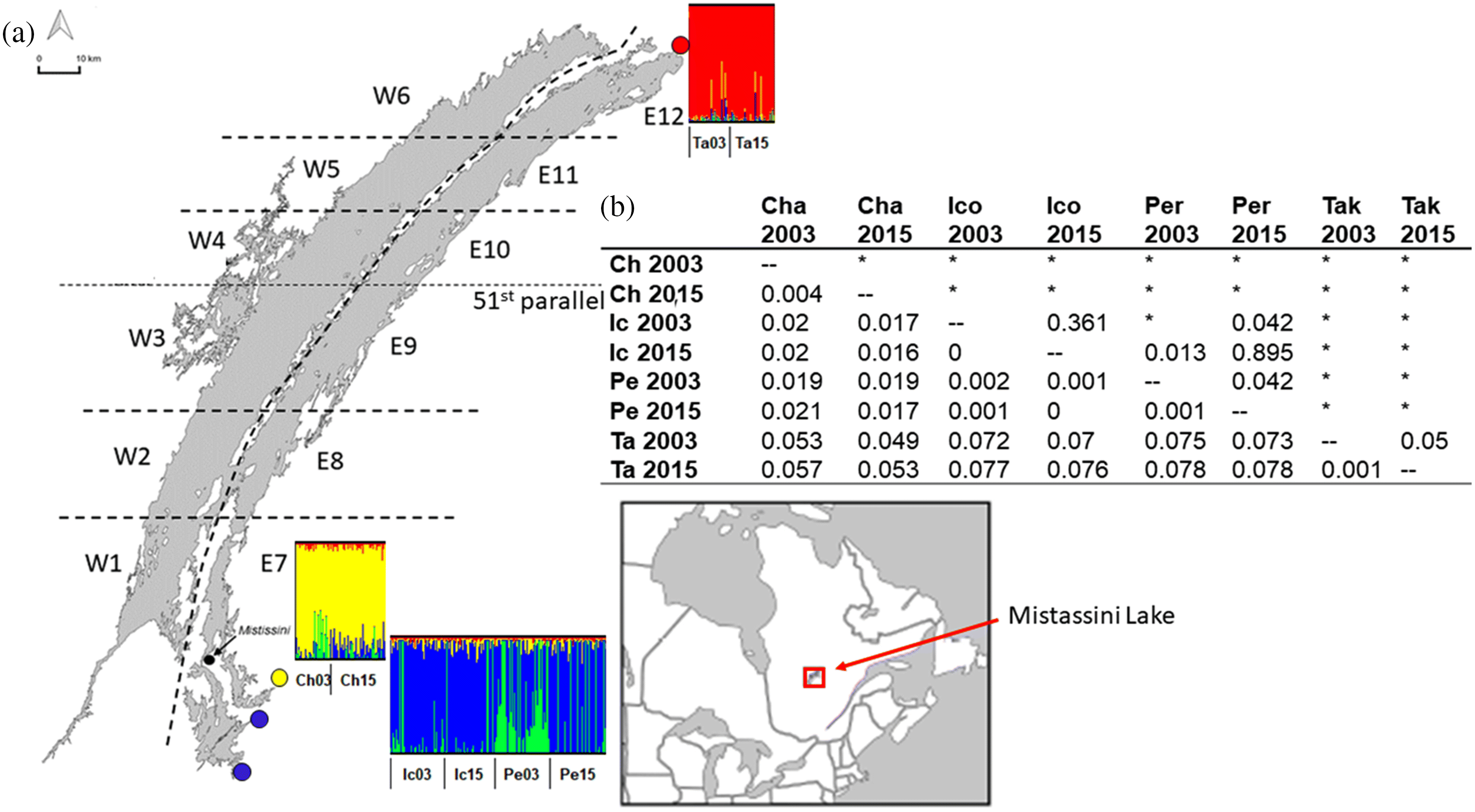
Population structure contrast
| Parameter | Species | Initial prediction | Result |
|---|---|---|---|
| Population structure/degree of philopatry/gene flow | walleye | Complementary | Congruent and complementary |
| Lake trout | Complementary | Complementary | |
| Pike | Complementary | Disagree | |
| Body size or morphology | Walleye | Complementary | Congruent and complementary |
| Lake trout | Congruent | Congruent and complementary | |
| Pike | Congruent | Congruent and complementary | |
| Conservation | Walleye | Complementary | Congruent for size, disagree for number of fish |
| Lake trout | Presented as complementary information | ||
| Pike | Presented as complementary information | ||
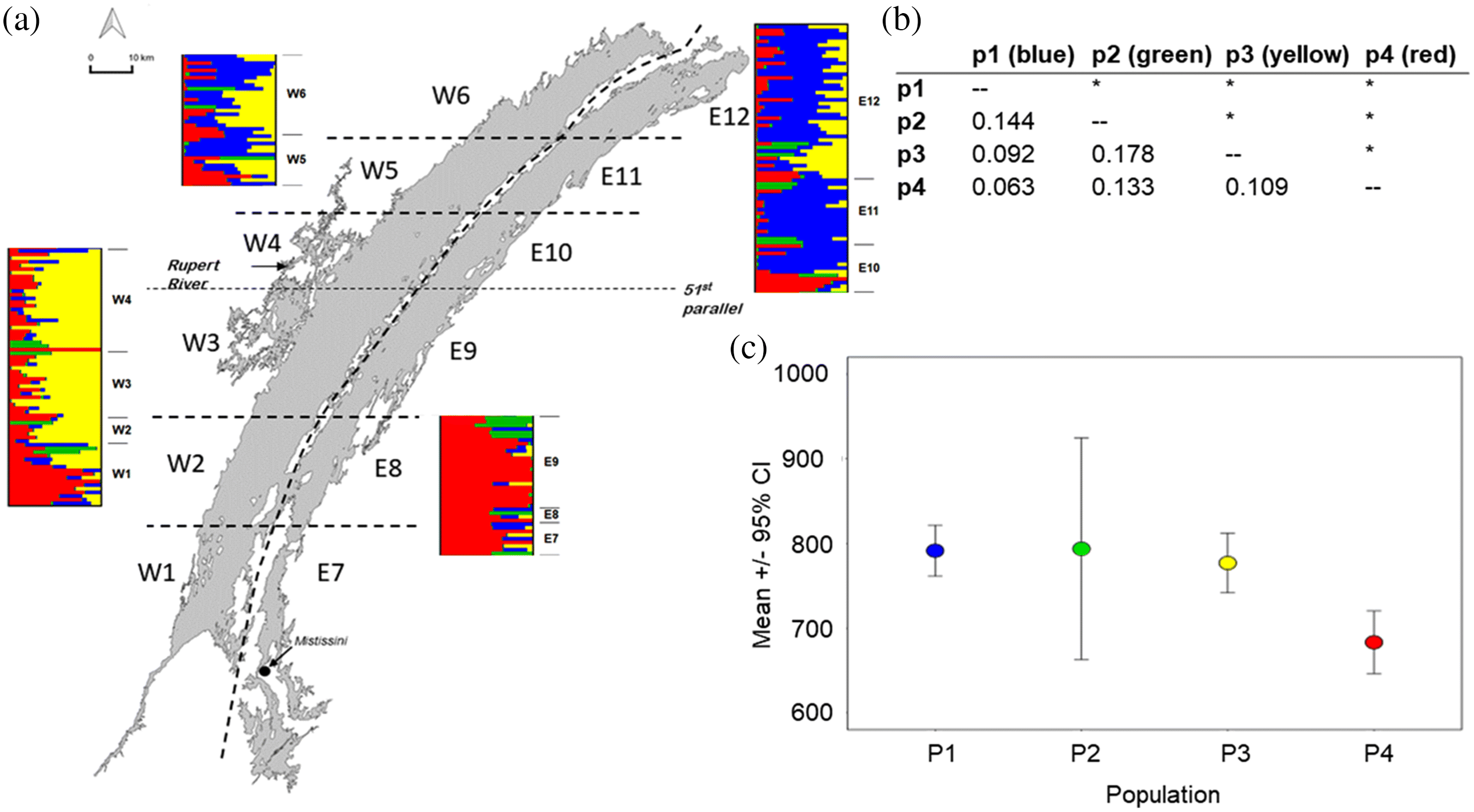
Population structure IEK
Population structure WSD
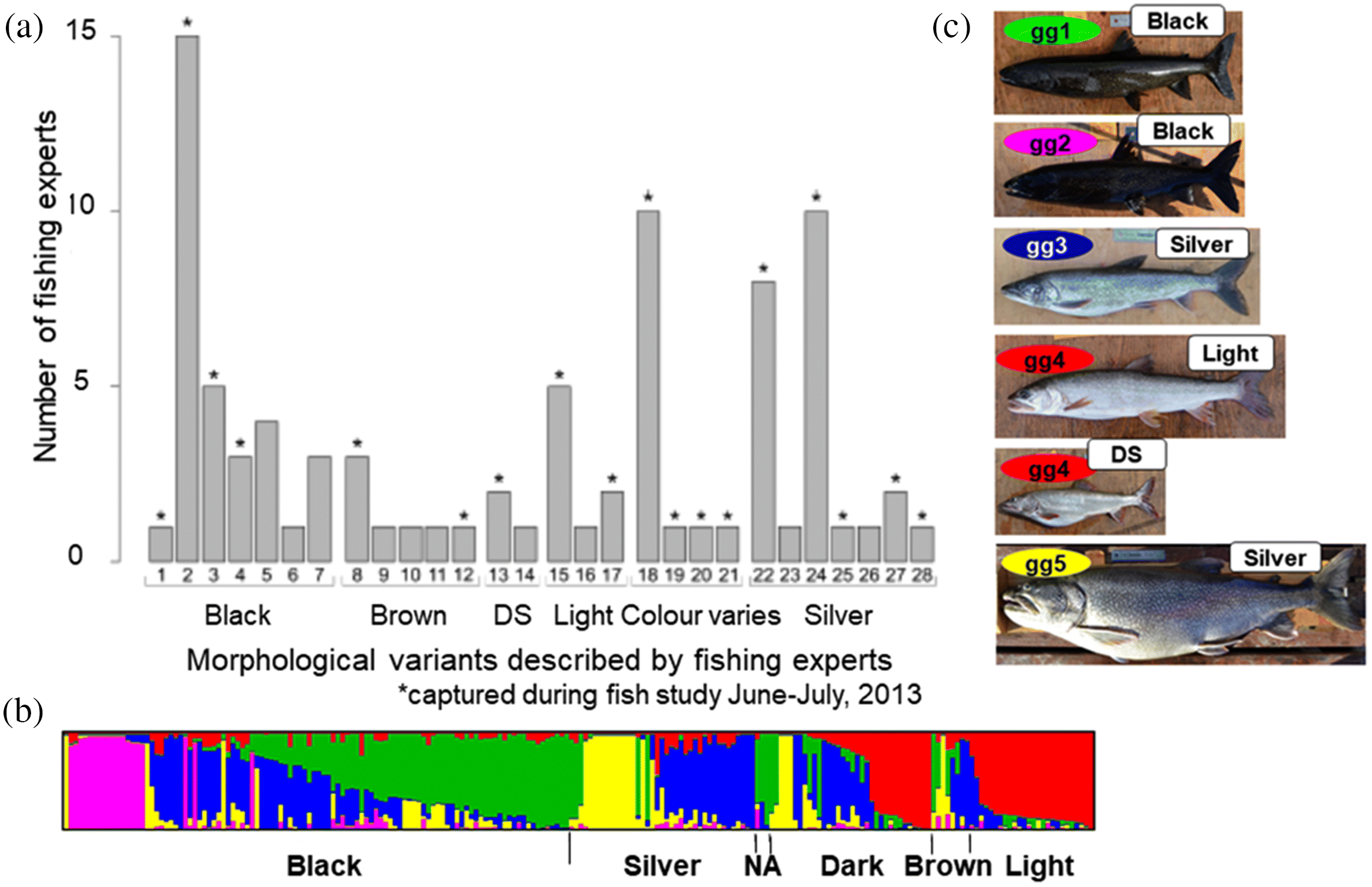
Morphology contrast and IEK outcomes
Morphology WSD
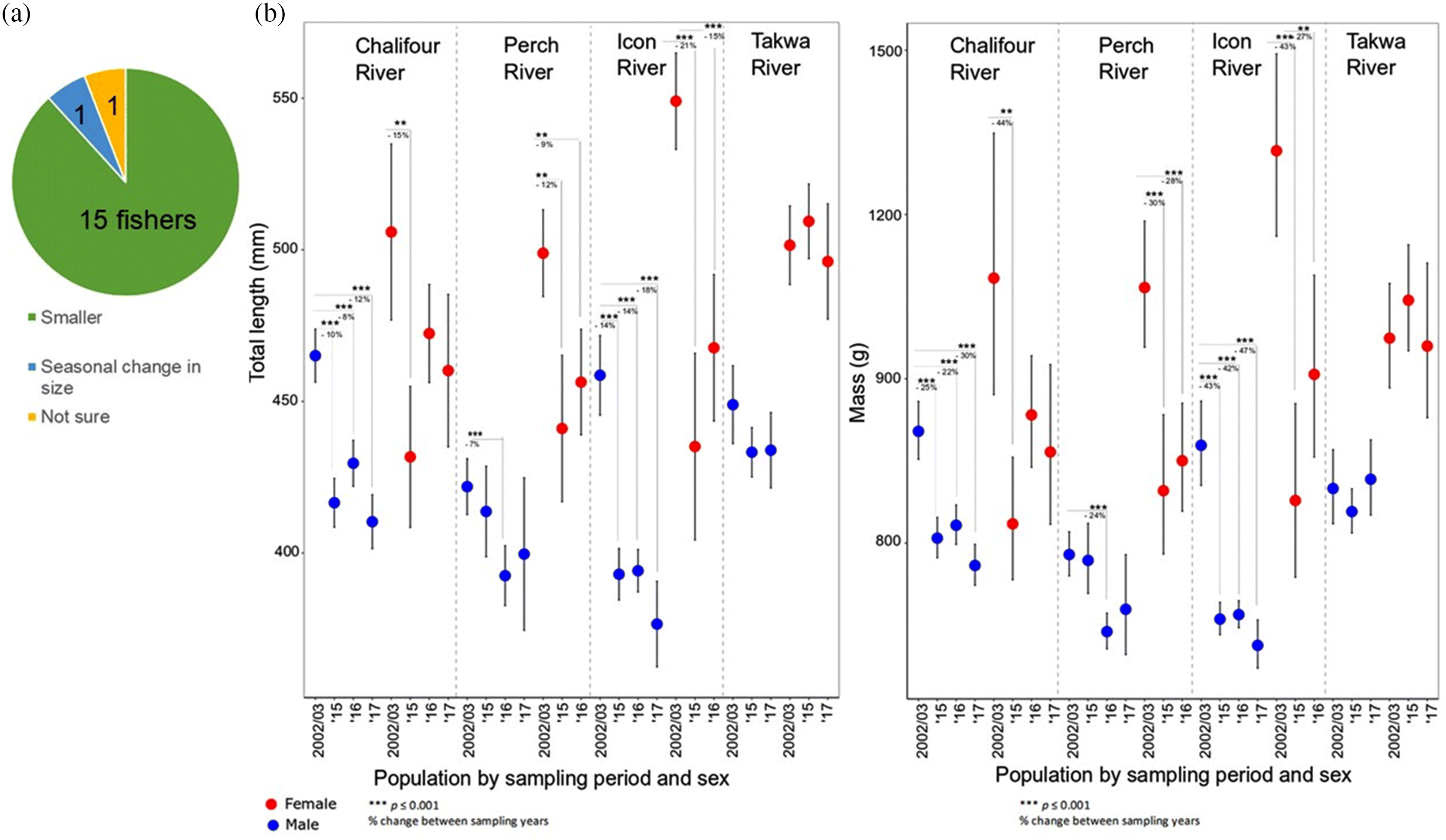
Conservation contrast
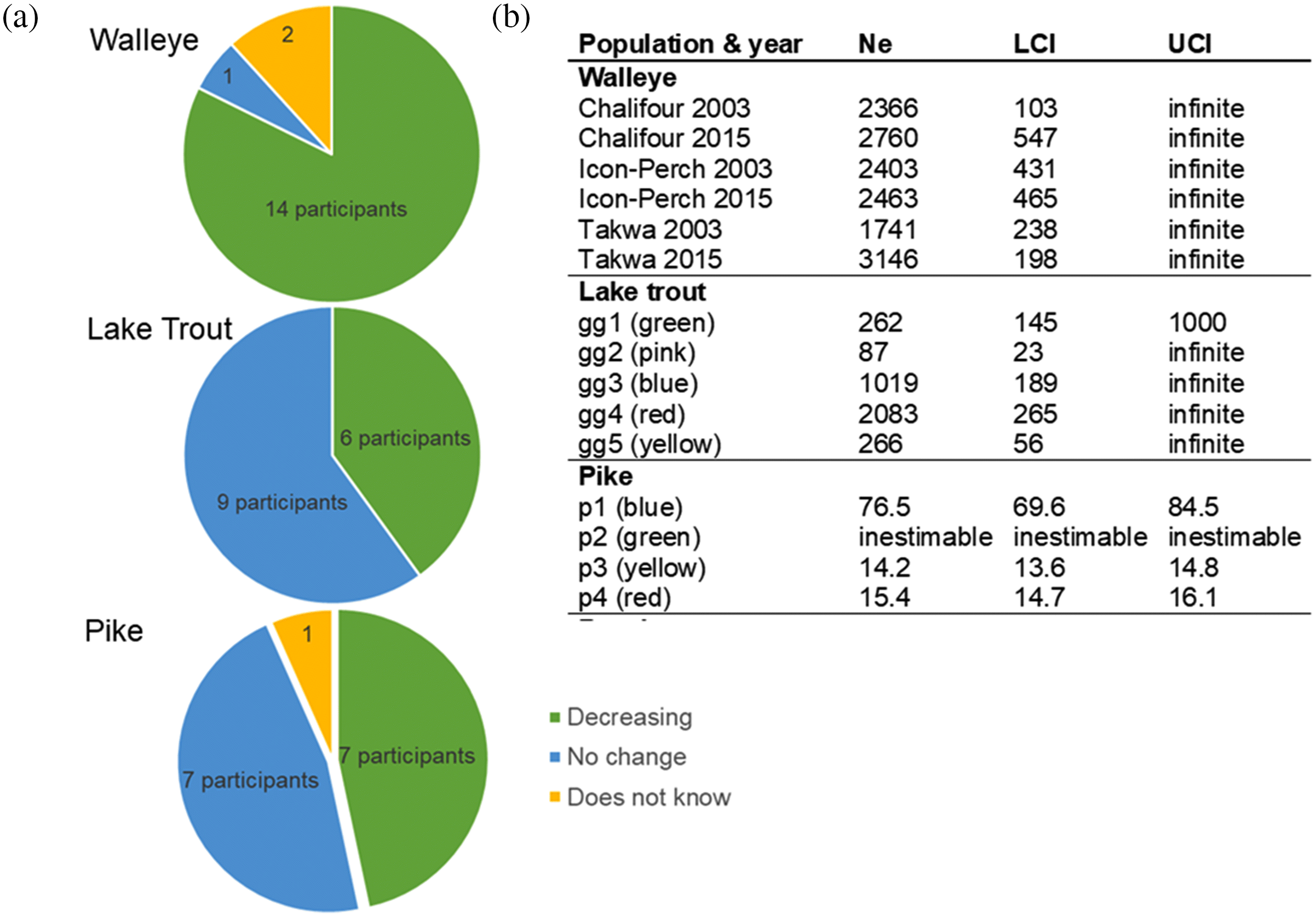
Conservation IEK
Conservation WSD
Lake trout
Population structure contrast
Population structure IEK
Population structure WSD
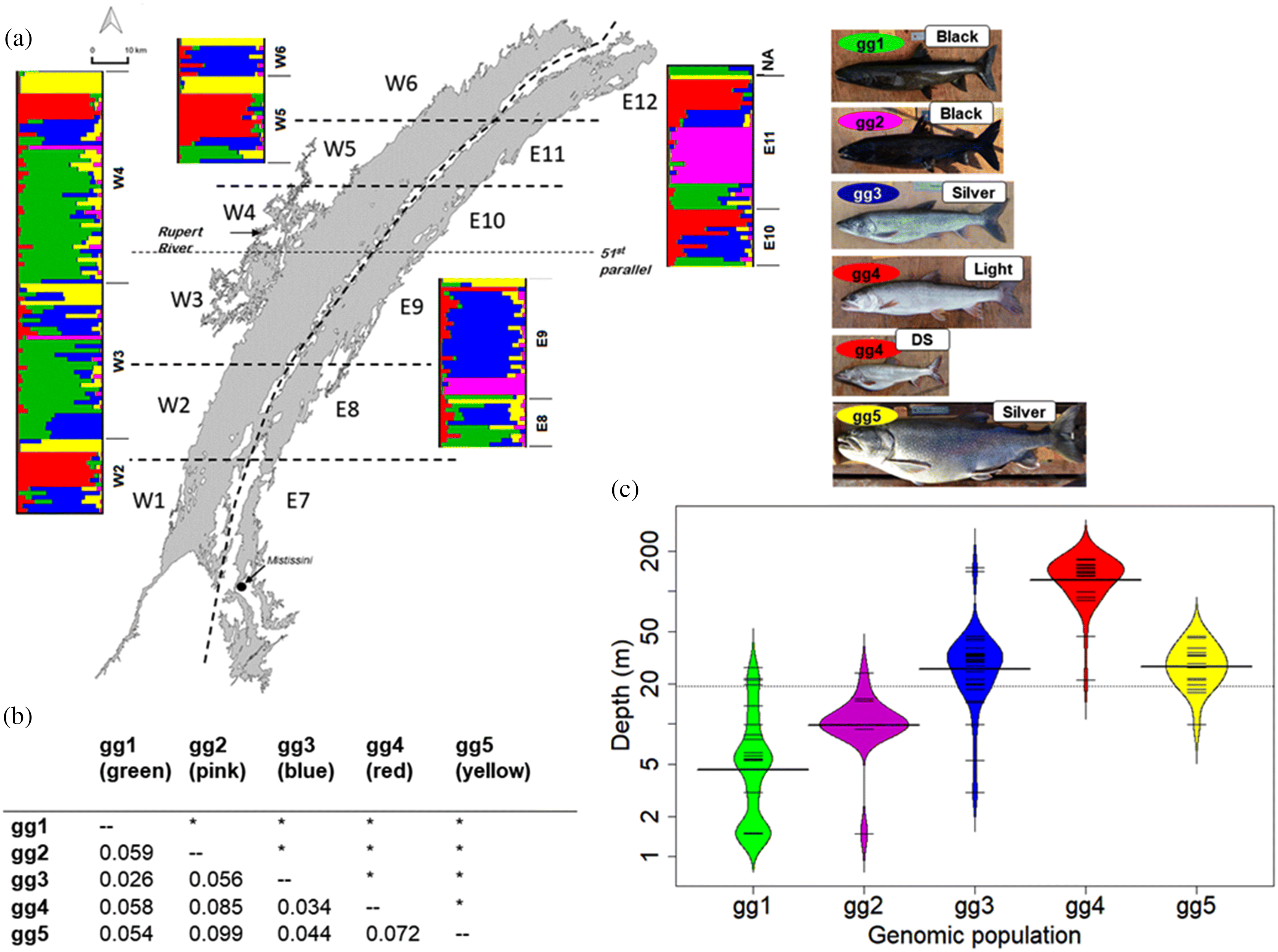
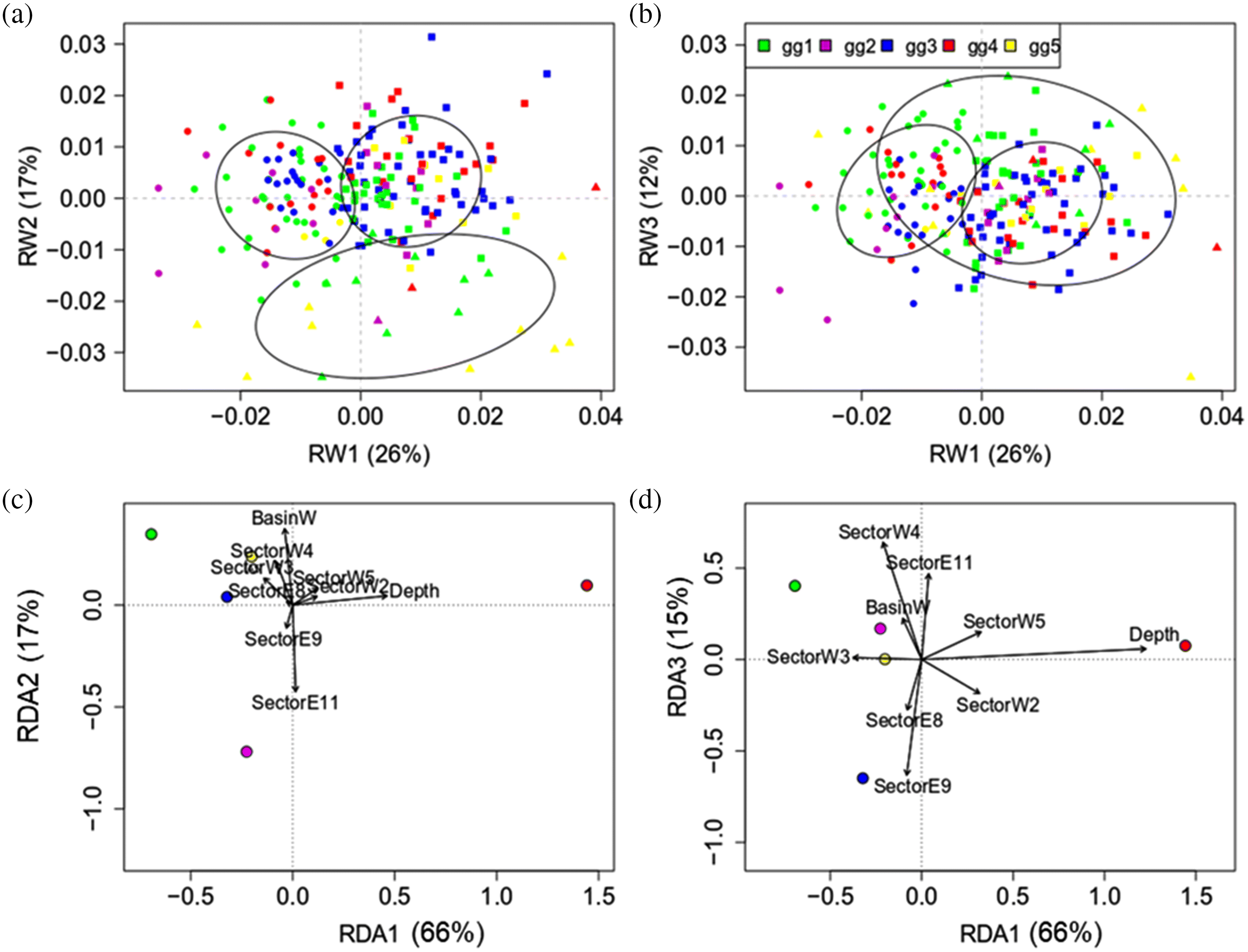
Morphology contrast
Morphology IEK and WSD
Conservation and population monitoring information
Conservation IEK
Conservation WSD
Northern pike
Population structure contrast
Population structure IEK
Population structure WSD
Morphology contrast, as well as IEK outcomes and WSD results
Conservation and population monitoring information
Conservation IEK
Conservation WSD
Discussion
Population structure—degree of philopatry
Morphological variation
Conservation status
Recommendations: Weaving IEK and WSD for freshwater fisheries monitoring
Positionality
Acknowledgements
Competing interest statement
References
Supplementary material
- Download
- 218.22 KB
Information & Authors
Information
Published In

History
Copyright
Data Availability Statement
Key Words
Sections
Subjects
Plain Language Summary
Authors
Author Contributions
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
There are no citations for this item
