SARS-CoV-2 detection from the built environment and wastewater and its use for hospital surveillance
Abstract
Patients hospitalized with SARS-CoV-2 infections are major contributors to morbidity and mortality in health care settings. Our understanding of the distribution of this virus in the built health care environment and wastewater, and relationship to disease burden, is limited. We performed a prospective multi-center study of environmental sampling of SARS-CoV-2 from hospital surfaces and wastewater and evaluated their relationships with regional and hospital COVID-19 burden. We validated a qPCR-based approach to surface sampling and collected swab samples weekly from different locations and surfaces across two tertiary care hospital campuses for a 10-week period during the pandemic, along with wastewater samples. Over the 10-week period, 963 swab samples were collected and analyzed. We found 61 (6%) swabs positive for SARS-CoV-2, with the majority of these (n = 51) originating from floor samples. Wards that actively managed patients with COVID-19 had the highest frequency of positive samples. Detection frequency in built environment swabs was significantly associated with active cases in the hospital throughout the study. Wastewater viral signal changes appeared to predate change in case burden. Our results indicate that environment sampling for SARS-CoV-2, in particular from floors, may offer a unique and resolved approach to surveillance of COVID-19.
Introduction
COVID-19 has caused substantial morbidity and mortality worldwide since it was first identified at the end of 2019. Despite the development of effective vaccines and the discovery of multiple therapeutic agents (Baden et al. 2021; Siemieniuk et al. 2020), the SARS-CoV-2 virus and agent of COVID-19 continues to transmit across all corners of the globe. As of November 2021, it is estimated that over 250 million people have been infected with SARS-CoV-2 from the start of the pandemic, with over 5 million deaths attributable to COVID-19 (World Health Organization 2021).
Hospitals were recognized early on as a potentially high-risk environment for acquiring COVID-19 infection for patients and health care workers (HCWs), with health care facility outbreaks and nosocomial acquisition of COVID-19 common and well described (Islam et al. 2020). The respiratory droplet route is considered to be the primary means for transmission of SARS-CoV-2 in hospitals; however, we still have a poor understanding of how the virus is distributed throughout the hospital physical environment and its implications both for surveillance and fomite-based transmission (Islam et al. 2020; Mondelli et al. 2021).
Laboratory experiments indicate that the virus can remain viable on stainless steel and plastics for approximately 6 h, and SARS-CoV-2 RNA can be detected from wastewater (van Doremalen et al. 2020). Preliminary studies have shown a range from no contamination to extensive contamination of hospital ward surfaces, including computer mice, trashcans, and door handles (Guo et al. 2020). In addition to identifying the potential routes for fomite-based transmission, surveying the distribution of SARS-CoV-2 throughout the hospital environment over time can provide insight into hospital burden and potential outbreak identification and monitoring. New approaches for early outbreak detection will be particularly important to reduce institutional morbidity and mortality (Smith et al. 2020). The built environment presence of SARS-CoV-2 could act as a spatially resolved indirect measure of symptomatic and asymptomatic carriage/infection within patients and health care workers (HCWs). In a similar fashion, wastewater surveillance may offer a less spatially resolved but potentially anticipatory mechanism for hospital COVID-19 surveillance and monitoring (Gonçalves et al. 2021; Westhaus et al. 2021).
In this study, we conducted a longitudinal assessment of the prevalence of SARS-CoV-2 on commonly contacted hospital surfaces, as well as from hospital wastewater, and we compared these markers with active COVID-19 case prevalence at two large tertiary care hospital campuses and in the surrounding community. We assessed the distribution of SARS-CoV-2 in the built environment by identifying locations and surface types where SARS-CoV-2 RNA was most prevalent. In addition, by testing the association between environmental detection and COVID-19 case prevalence, we examined the potential use of built environmental detection as a surveillance tool for predicting active case burdens.
Materials and methods
SARS-CoV-2 detection from swab samples
Surfaces were sampled with the P-208 Environmental Surface Collection Prototype kit from DNA Genotek, consisting of a flocked swab and a semi-lytic nucleic acid stabilization solution for post-collection swab immersion. SARS-CoV-2 was detected by quantitative reverse-transcriptase polymerase chain reaction (qPCR) of RNA extracted from sample stabilization solution using the MagMAX Viral/Pathogen II (MVP II) Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA). Extractions were performed in batches of up to 96 in deep-well plates according to the manufacturer’s protocol, with reagent volumes modified proportionally for a 300 μL sample input volume, and an elution volume of 50 μL. Each batch included positive (heat-inactivated SARS-CoV-2 virus) and negative (stabilization solution only) control samples to monitor extraction efficacy and potential cross-contamination. None of the control samples yielded false positive or false negative detections.
qPCR was performed with primers and a TaqMan probe targeting the N1 region of the SARS-CoV-2 nucleocapsid gene (Centers for Disease Control and Prevention 2020) (Supplementary Material 1, Table S1). Each qPCR reaction consisted of 5 μL of extracted RNA, 300 nM of each primer, 200 nM of probe, and 5 μL of 4× TaqPath 1-Step Multiplex Master Mix (No ROX) (Thermo Fisher Scientific, Waltham, MA) in a final volume of 20 μL. Thermal cycling, fluorescence quantification, and quantitative cycle (Cq) determination were performed on a Bio-Rad CFX Connect with the following cycling conditions: 25 °C for 2 min, 50 °C for 15 min, 95 °C for 2 min, and 45 cycles of denaturation (95 °C for 3 s) and elongation (55 °C for 30 s).
Limit of detection analysis
Limits of detection (LOD) and quantification (LOQ) were determined by analyzing 10-fold serial dilutions of SARS-CoV-2 RNA (ATCC VR-1986D; lot 70035624; 4.73 × 103 genome copies/μL) and heat-inactivated virus (ATCC VR-1986HK; lot 70035039; 3.75 × 105 genome copies/μL) standards obtained from Cedarlane Laboratories (Burlington, ON). Dilutions of the SARS-CoV-2 RNA standard were analyzed by qPCR with 14–20 replicates per dilution. For the virus standard, RNA was extracted from serial dilutions of heat-inactivated virus (7 replicate extractions at each dilution), followed by qPCR analysis.
LODs and LOQs were estimated using a LOD Calculator (Klymus et al. 2020) (R script available at: github.com/cmerkes/qPCR_LOD_Calc). The standard curves were modeled as linear regressions based on the middle two quartiles of Cq values for each standard concentration exhibiting >50% detection. The LOD was defined as the lowest input amount that was detected in >95% of replicates, and the LOQ was defined as the lowest input amount yielding reproducible Cq values with a coefficient of variation less than 0.35. The LOD and LOQ of the RNA standards were estimated by a curve-fitting approach (Klymus et al. 2020), while the LOD of the heat-inactivated virus was determined using a discrete threshold (i.e., the lowest tested input amount with >95% detection).
Swabbing method and validation
The swabbing method was validated in laboratory experiments measuring recovery of heat-inactivated SARS-CoV-2 virus from sheets of acrylic, stainless steel, and vinyl upholstery. Each material was spiked in triplicate with 105 virus copies, pipetted as 30–40 small spots within 2.25″ × 2.25″ squares. After a 2.5 h drying period, swabs were prewetted with stabilization solution, surfaces were swabbed for 30 s, and swabs were stored in stabilization solution in collection vials. Control samples consisted of virus spiked directly into collection vials with stabilization solution and unused swabs. Following one day of storage at room temperature, RNA was extracted and SARS-CoV-2 RNA was detected by qPCR. Virus genome copy numbers were estimated using the virus standard curve that relates Cq values to input genome copies. The percent virus recovery was determined by dividing the amount of virus estimated for the surface swabbed samples by the amount of virus estimated in the control samples. Significant differences in recovery efficiencies across surfaces were determined by ANOVA and Tukey’s honestly significant difference test.
Built environment sample collection
We prospectively collected built environment swabs from two large tertiary-care, academic hospital campuses (Campus A and Campus B, totaling over 1100 beds) from Ottawa, Ontario, Canada, over a 10-week period between 28 September and 6 December 2020. We obtained between 40 and 50 samples from each campus weekly, generally maintaining location (ward/unit) and surface type longitudinally; however, we did not attempt to sample the exact same item/spot longitudinally or coordinate with cleaning schedules to improve our study generalizability. For example, a computer keyboard sampled in week 1 on Unit A, may be different from a computer keyboard sampled on Unit A in week 2. At both campuses, we collected samples from two COVID-dedicated noncritical care units, two intensive care (ICU) COVID-designated units, two emergency department (ED) areas, non-COVID medical and surgical units (n = 3 at Campus A, n = 4 at Campus B), one ambulatory unit (dialysis), public/nonpatient-care spaces within the hospital (e.g., main elevators), and exterior hospital grounds (e.g., parking garage). A designated HCW space was also sampled at Campus A. Built environment samples were collected by wetting a P-208 kit flocked swab with the stabilization media and sweeping the swab tip across an approximately 2″ × 2″ area (dependent upon the item/surface size) for 30 s. The swab was then transported in the P-208 stabilization media and stored at ambient temperature to be processed/analyzed within 1 month of collection. A run-in week was performed prior to week 1, to test sample collection, logistics, and sample processing, but is not included in this analysis as a number of specimen sites were initially not included for this week, making inter-week comparisons challenging.
Specific (noncritical care) wards in the hospital were designated for the ongoing inpatient care of COVID-19 patients, and these were designated as “COVID wards”. However, we classified both the COVID Wards as well as the ICU and ED as “COVID risk units”, because they were anticipated to be managing many SARS-CoV-2 infected patients.
Hospital practices
The visitor policy at the start of the study period limited visitation to one visitor per patient per day; however, at the start of study week 4, visitors were further restricted to essential caregivers only. Environmental cleaning was in keeping with provincial best practice and occurred once daily in both public and clinical areas sampled, with all contact points disinfected with Vert-2-Go everyday disinfectant (quaternary-ammonium based product) and floors mopped with Vert-2-Go Oxy floor cleaner (hydrogen peroxide based product). The only deviation from this was once weekly cleaning of the Campus A parking garage until sometime between study weeks 5 and 7, then once daily thereafter. Infection Prevention and Control (IPAC) policies were also consistent with provincial best practice (Public Health Ontario 2020; van Doremalen et al. 2020). Throughout the study period, all staff, visitors, and patients were screened for COVID-19 symptoms and risk factors (e.g., exposures, international travel) on entry to the hospital, in accordance with Ontario Ministry of Health (2020a) guidance. Universal masking for all staff and visitors was introduced prior to the study period. Universal eye protection (visor or goggles) for staff working in clinical areas was introduced at the start of study week 1.
Wastewater sample collection
Raw sewage grab samples from Campus A were taken with a 5 L bucket on a rope between 11:10 am and 12:30 pm each sampling day, which occurred once every 1 to 2 weeks during (and immediately prior to) the study period. The samples were swirled in the bucket and quickly poured into prelabelled sterile 1 L Nalgene bottles which were immediately placed into a cooler with ice packs for transport back to the lab and processed within 1 hr. Temperature of the samples, measured at the time of sampling with an infrared thermometer, for October–December were between 12 and 22 °C. A 200 mL aliquot was concentrated to 0.25–1.25 mL using the Centricon Plus-70 (MilliporeSigma, Burlington, MA) centrifugal ultrafilter with a cut-off of 10 kDa (Medema et al. 2020). The resulting concentrate was extracted using the RNeasy PowerMicrobiome Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions with an elution of 100μL and SARS-CoV-2 RNA was quantified by qPCR.
Real-time quantitative PCR analysis of wastewater samples
Primers and probes, and real-time reverse transcriptase (RT RT-PCR) methods developed by the US Centers for Disease Control and Prevention (2020) were used with the Bio-Rad CFX96 Touch Real-Time PCR Detection System. The RT RT-PCR thermal cycling protocol used to amplify the N1 and N2 target regions within the SARS-CoV-2 nucleocapsid gene, was carried out at 50 °C for 30 min, followed by 95 °C for 15 min and 50 cycles of 95 °C for 15 and 60 °C for 30 s. Reaction mixtures for RT RT-PCR included 5 μL RNA template, 4 μL of QIAGEN OneStep RT-PCR 5 × Buffer, 2 μL dNTP mix (10 mM each), 2 μL Enzyme Mix, Primers and Probes (500 nM each), and 2 μL of 4 mg/mL bovine serum albumen. Each reaction was adjusted to a final volume of 20 μL with RNase-free water. Each sample was run in duplicate. An MS2 bacteriophage matrix spike whole process control was used to qualitatively assess the efficiency of the concentration process and RNA extraction as well as confirm RT RT-PCR protocols. RT RT-PCR inhibition control using MS2 bacteriophage spiked into raw sewage RNA extracts and molecular grade water was used to assess the performance of the RNA extraction, RT-PCR, and to detect the presence of inhibitors. Quantification of the N1 and N2 gene targets in the raw sewage sample concentrates was done using standard curves created with a 2019-nCoV_N_Positive Control plasmid (Integrated DNA Technologies, Coralville, IA) Nontemplate controls were run for each mastermix preparation with molecular grade water as the template. For each week under observation, we averaged the detectable copies per millilitre from both N1 and N2 gene targets.
Hospital and citywide COVID-19 cases and outbreaks
Daily active patient COVID-19 case counts and locations, including admitted and nonadmitted ED cases, were obtained from IPAC. Cases were those confirmed by RT-PCR. We generated a measure of weekly “active” COVID-19 cases by taking the mean of these daily active cases for each week (typically data only available for weekdays). HCW cases were not included in the analysis, as HCWs were more likely than patients to spend time on multiple units and move throughout the hospital. Also, once a HCW became symptomatic or had a positive COVID-19 test result, they would no longer remain within the hospital. Hospital outbreak data were obtained in real time from IPAC and verified post-study on the publicly available Ottawa Public Health (OPH) COVID-19 dashboard (Ottawa Public Health 2021). An outbreak was defined as per the Ontario Ministry of Health (2020b) as, “Two or more laboratory-confirmed COVID-19 cases (patients and (or) staff) within a specified area (unit/floor/service) within a 14-day period where both cases could have reasonably acquired their infection in the hospital”. New and total active COVID-19 cases in Ottawa were obtained daily from the OPH COVID-19 dashboard, and a weekly “active” caseload was calculated as the mean of the daily active cases across a given week.
Data analysis
To evaluate the characteristics associated with positive swab results, we performed a logistic regression model using specimen characteristics as predictors of built environment sample positivity (total or floor specific specimens). Specimen characteristics were coded as categorical variables, including unit COVID risk type, object/site, material, and study week. The outcome (sample positivity) was coded as a binary variable (1 = detected, 0 = not-detected).
We also evaluated the strength of associations between (i) proportion of hospital built environment swabs positive for SARS-CoV-2 or (ii) proportion of hospital floor swabs positive for SARS-CoV-2 and hospitalized active COVID-19 cases. We used a negative-binomial regression model with active cases as the outcome. We did not quantitatively evaluate the association between wastewater and active cases because of the limited number of wastewater data points.
For the majority of the analyses we considered the two campuses in combination, as they reflect a single health care center with transfer occurring within and between units in both campuses. We also chose not to evaluate case burden by specific unit because, in general, as soon as a case was identified on a given non-COVID risk unit, the patient would be transferred to a specific COVID Risk unit either for cohorting or due to severity of illness (e.g., ICU).
All statistical analyses were performed using R (Version 4.0.3). Values used to build the figures are in the datasets found in Supplementary Material 2. Research ethics board approval was obtained through the Ottawa Health Science Network Research Ethics Board for this study.
Results
Limit of detection and swab validation
SARS-CoV-2 RNA was detected and quantified by qPCR analysis using the N1 primer and probe set targeting the nucleocapsid gene (Centers for Disease Control and Prevention 2020). We analyzed a dilution series of a SARS-CoV-2 RNA standard to produce a standard curve relating the Cq values to genome copy number and determine the LOD and LOQ (Fig. 1a). We estimated that reactions containing at least 3 genome copies were reliably detected under our qPCR conditions (detection rate greater than 95%), whereas precise quantification required at least 150 copies (Cq < 30) (Fig. 1a).
Fig. 1.
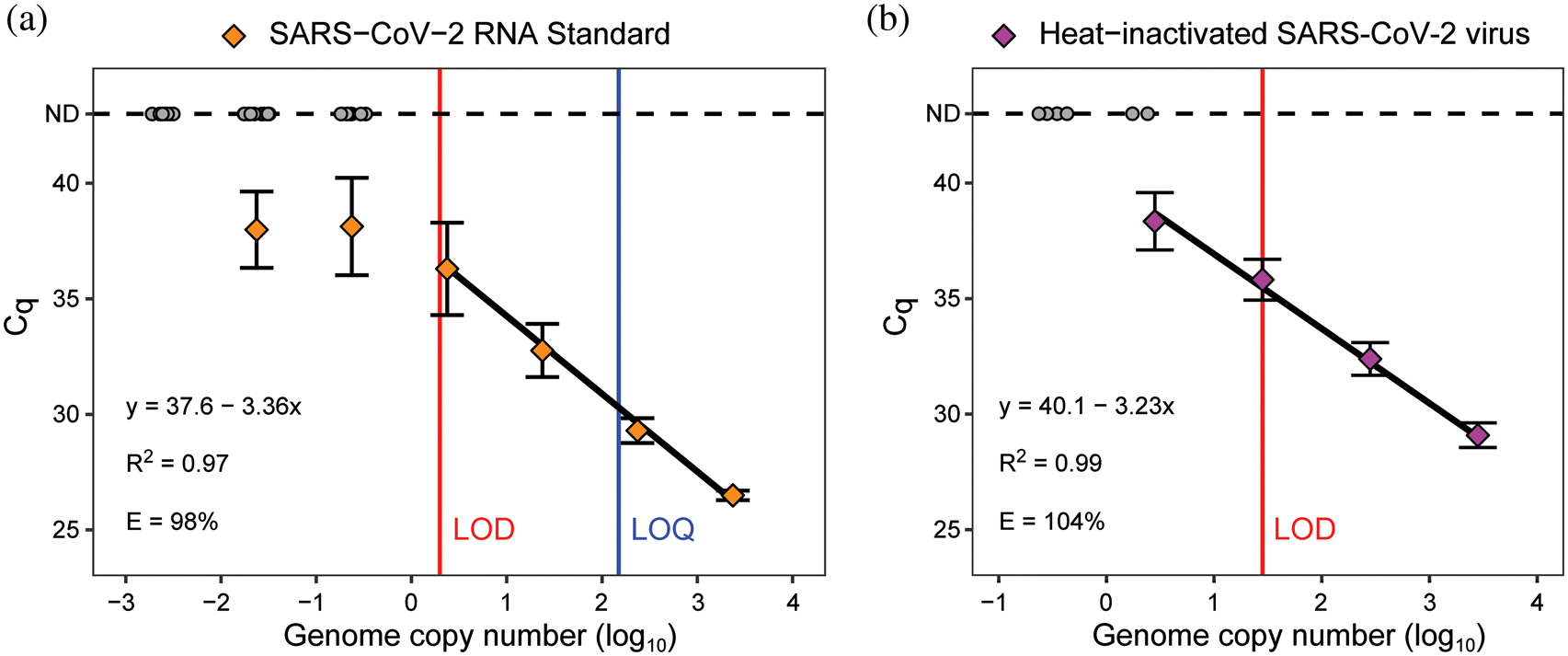
Analysis of the built environment swab samples involved a nucleic acid extraction step prior to qPCR analysis. We evaluated the extraction method by qPCR analysis of RNA extracted from samples of a heat-inactivated SARS-CoV-2 virus standard (Fig. 1b). The LOD for the virus standard (28 copies per reaction) was ∼10-fold higher than that of the RNA standard, which can be attributed to RNA loss during the extraction procedure, presence of PCR inhibitors in the eluant, or inaccuracies in the reported titers of the standards. Nevertheless, the procedure was sensitive and reproducible, and the results yielded a linear standard curve for converting Cq values to viral copies while accounting for inaccuracies introduced by sample processing. We validated the swabbing method by estimating the recovery of virus from surfaces spiked with heat-inactivated SARS-CoV-2 (Supplementary Material 1, Fig. S1). The mean recovery efficiencies ranged from 28% to 42% across three different surface types, which are values similar to those reported in a previous study of SARS-CoV-2 surface swabbing (Parker et al. 2020).
SARS-CoV-2 RNA detection in wastewater
Standard curves generated for both N1 and N2 targets gave consistent results with PCR efficiencies between 90% and 110%, and R2 values greater than 0.99. Nontemplate controls were negative for all runs. There was a shift in sensitivity from the N2 target showing lower Cq value than N1 for the first 3 weeks followed by lower Cq values for N1 for the remainder of the study period. As a result of this, the data are presented as an average of N1 and N2 gene copy numbers.
Built environment screening
Over a 10-week period from 28 September to 6 December 2020 we systematically performed 963 built environment swabs across two tertiary care hospital sites. SARS-CoV-2 was detected from 6% of swabs (61/963), and it was commonly found from floor samples, with a detection prevalence on floors of 27% (51/188). SARS-CoV-2 was less frequently identified from elevator buttons, benches, items (e.g., hand sanitizer pumps, equipment carts), telephones, and computer keyboards.
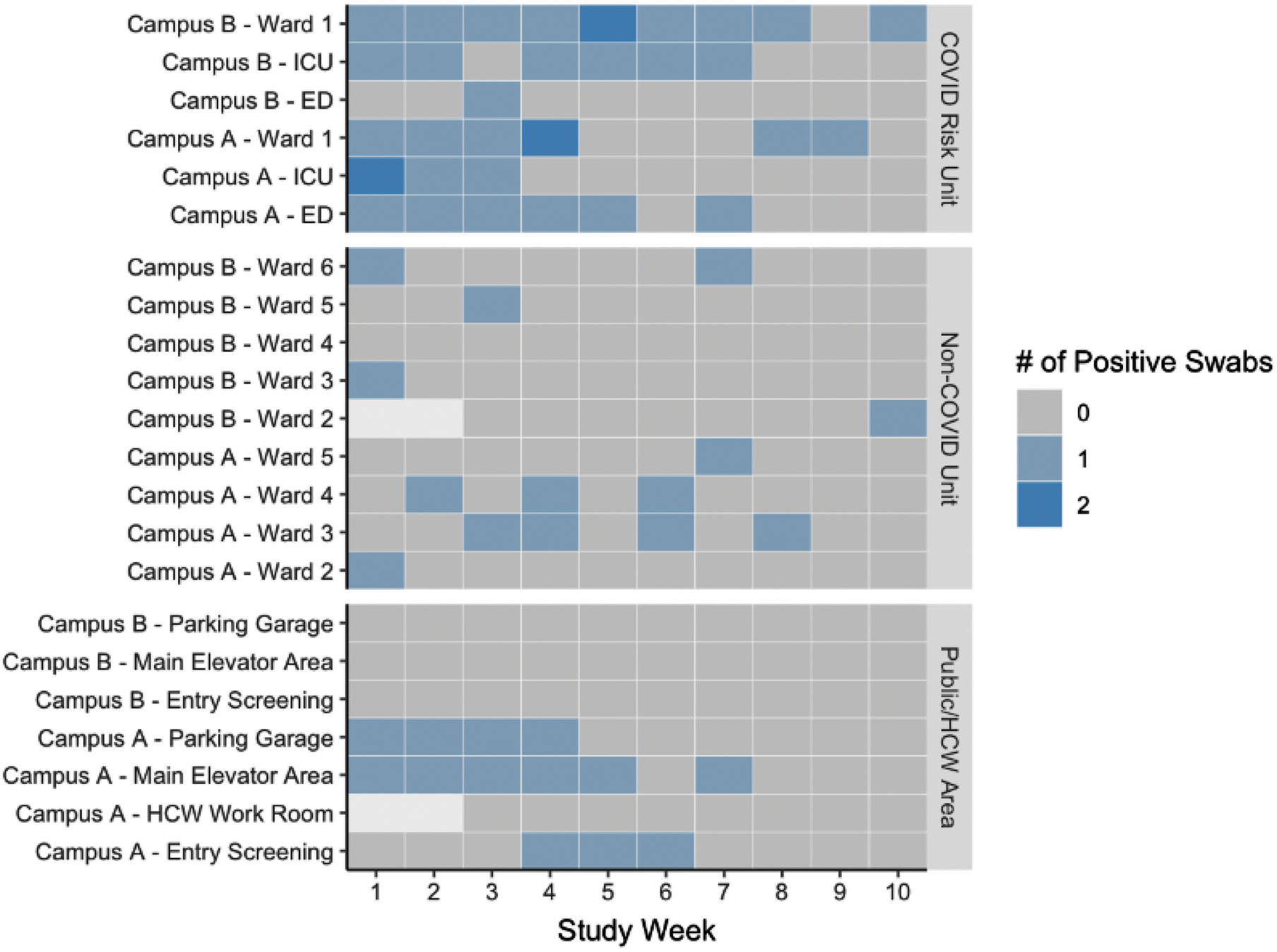
COVID risk units, namely COVID treatment wards, ICUs, and EDs, were the most common locations to find SARS-CoV-2 in the hospital environment, with detection prevalence of 18% (17/92), 8% (10/118), and 7% (7/100), respectively (Table 1 and Fig. 2). Non-COVID and public or HCW spaces tended to have a low frequency of SARS-CoV-2 detection from the environment. During this study, there were two outbreaks involving study units, at Campus B in the ED (one patient and two HCWs; study week 2–4) and at Campus A on Ward 3 (five patients and one HCW; study week 4–8). A third outbreak occurred on a non-COVID inpatient unit at Campus A, but this unit was not sampled in the study.
Table 1.
| Campus A | Campus B | Total | ||||
|---|---|---|---|---|---|---|
| Detected (N = 39)a | Not detected (N = 451) | Detected (N = 22) | Not detected (N = 451) | Detected (N = 61) | Not detected (N = 902) | |
| Unit COVID risk type | ||||||
| COVID risk unit | 17 (43.6%) | 150 (33.3%) | 17 (77.3%) | 126 (27.9%) | 34 (55.7%) | 276 (30.6%) |
| Non-COVID unit | 9 (23.1%) | 221 (49.0%) | 5 (22.7%) | 266 (59.0%) | 14 (23.0%) | 487 (54.0%) |
| Public/health care worker area | 13 (33.3%) | 80 (17.7%) | 0 (0%) | 59 (13.1%) | 13 (21.3%) | 139 (15.4%) |
| Object/site | ||||||
| Air (control) | 0 (0%) | 10 (2.2%) | 0 (0%) | 10 (2.2%) | 0 (0%) | 20 (2.2%) |
| Computer keyboard | 0 (0%) | 75 (16.6%) | 1 (4.5%) | 77 (17.1%) | 1 (1.6%) | 152 (16.9%) |
| Door handle | 0 (0% | 88 (19.5%) | 0 (0%) | 97 (21.5%) | 0 (0%) | 185 (20.5%) |
| Elevator buttons | 4 (10.3%) | 46 (10.2%) | 2 (9.1%) | 57 (12.6%) | 6 (9.8%) | 103 (11.4%) |
| Exterior bench | 1 (2.6%) | 9 (2.0%) | 0 (0%) | 0 (0%) | 1 (1.6%) | 9 (1.0%) |
| Floor | 32 (82.1%) | 59 (13.1%) | 19 (86.4%) | 78 (17.3%) | 51 (83.6%) | 137 (15.2%) |
| Hand sanitizer | 1 (2.6%) | 49 (10.9%) | 0 (0%) | 59 (13.1%) | 1 (1.6%) | 108 (12.0%) |
| Other | 1 (2.6%) | 37 (8.2%) | 0 (0%) | 10 (2.2%) | 1 (1.6%) | 47 (5.2%) |
| Table | 0 (0%) | 3 (0.7%) | 0 (0%) | 2 (0.4%) | 0 (0%) | 5 (0.6%) |
| Telephone receiver | 0 (0%) | 75 (16.6%) | 0 (0%) | 61 (13.5%) | 0 (0%) | 136 (15.1%) |
| Material | ||||||
| Air (Control) | 0 (0%) | 10 (2.2%) | 0 (0%) | 9 (2.0%) | 0 (0%) | 19 (2.1%) |
| Metal | 2 (5.1%) | 120 (26.6%) | 2 (9.1%) | 154 (34.1%) | 4 (6.6%) | 274 (30.4%) |
| Plastic/vinyl | 37 (94.9%) | 318 (70.5%) | 20 (90.9%) | 287 (63.6%) | 57 (93.4%) | 605 (67.1%) |
| Wood | 0 (0%) | 3 (0.7%) | 0 (0%) | 1 (0.2%) | 0 (0%) | 4 (0.4%) |
| Study Week | ||||||
| 1 | 7 (17.9%) | 41 (9.1%) | 4 (18.2%) | 44 (9.8%) | 11 (18.0%) | 85 (9.4%) |
| 2 | 6 (15.4%) | 43 (9.5%) | 2 (9.1%) | 45 (10.0%) | 8 (13.1%) | 88 (9.8%) |
| 3 | 6 (15.4%) | 40 (8.9%) | 3 (13.6%) | 45 (10.0%) | 9 (14.8%) | 85 (9.4%) |
| 4 | 8 (20.5%) | 39 (8.6%) | 2 (9.1%) | 46 (10.2%) | 10 (16.4%) | 85 (9.4%) |
| 5 | 3 (7.7%) | 47 (10.4%) | 3 (13.6%) | 43 (9.5%) | 6 (9.8%) | 90 (10.0%) |
| 6 | 3 (7.7%) | 47 (10.4%) | 2 (9.1%) | 44 (9.8%) | 5 (8.2%) | 91 (10.1%) |
| 7 | 3 (7.7%) | 47 (10.4%) | 3 (13.6%) | 45 (10.0%) | 6 (9.8%) | 92 (10.2%) |
| 8 | 2 (5.1%) | 48 (10.6%) | 1 (4.5%) | 46 (10.2%) | 3 (4.9%) | 94 (10.4%) |
| 9 | 1(2.6%) | 49 (10.9%) | 0 (0%) | 47 (10.4%) | 1 (1.6%) | 96 (10.6%) |
| 10 | 0 (0%) | 50 (11.1%) | 2 (9.1%) | 46 (10.2%) | 2 (3.3%) | 96 (10.6%) |
a
The number and percentage of positive (or negative) detections associated with each sample location or characteristic are given. For example, 43.6% of all positive detections (17/39) in Campus A were associated with samples collected from COVID risk units.
Fig. 2.
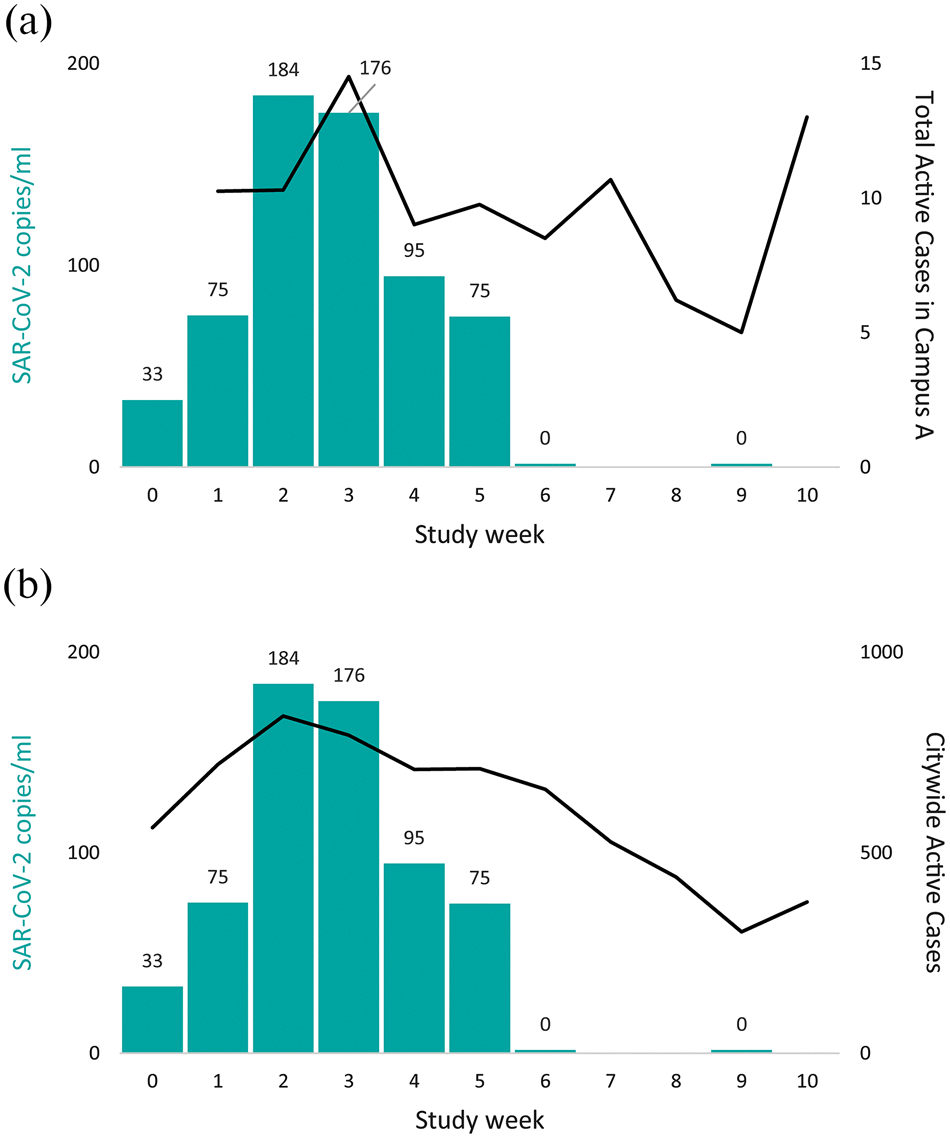
Across the 10-week study period, SARS-CoV-2 detection prevalence paralleled total active cases across the two hospital campuses. Viral prevalence gradually declined over time, but with spikes in hospitalized cases being clearly reflected in built environment burden (Fig. 3a), as well as an increase at week 10 reflecting the start of hospitalizations in the second wave of the pandemic within the city. Built environment burden also paralleled citywide cases, but with less concordance (Fig. 3b). Campus B tended to have SARS-CoV-2 positivity largely restricted to COVID risk units, but Campus A seemed to have more activity in non-COVID risk units/public spaces. A multi-variable logistic regression model identified that increasing study week was associated significantly with reduced detection of SARS-CoV-2 (p < 0.05), and that COVID Risk units, Campus A, and some specific materials and objects were significantly associated with increased SARS-CoV-2 detection (Table 1 and Table 2).
Fig. 3.
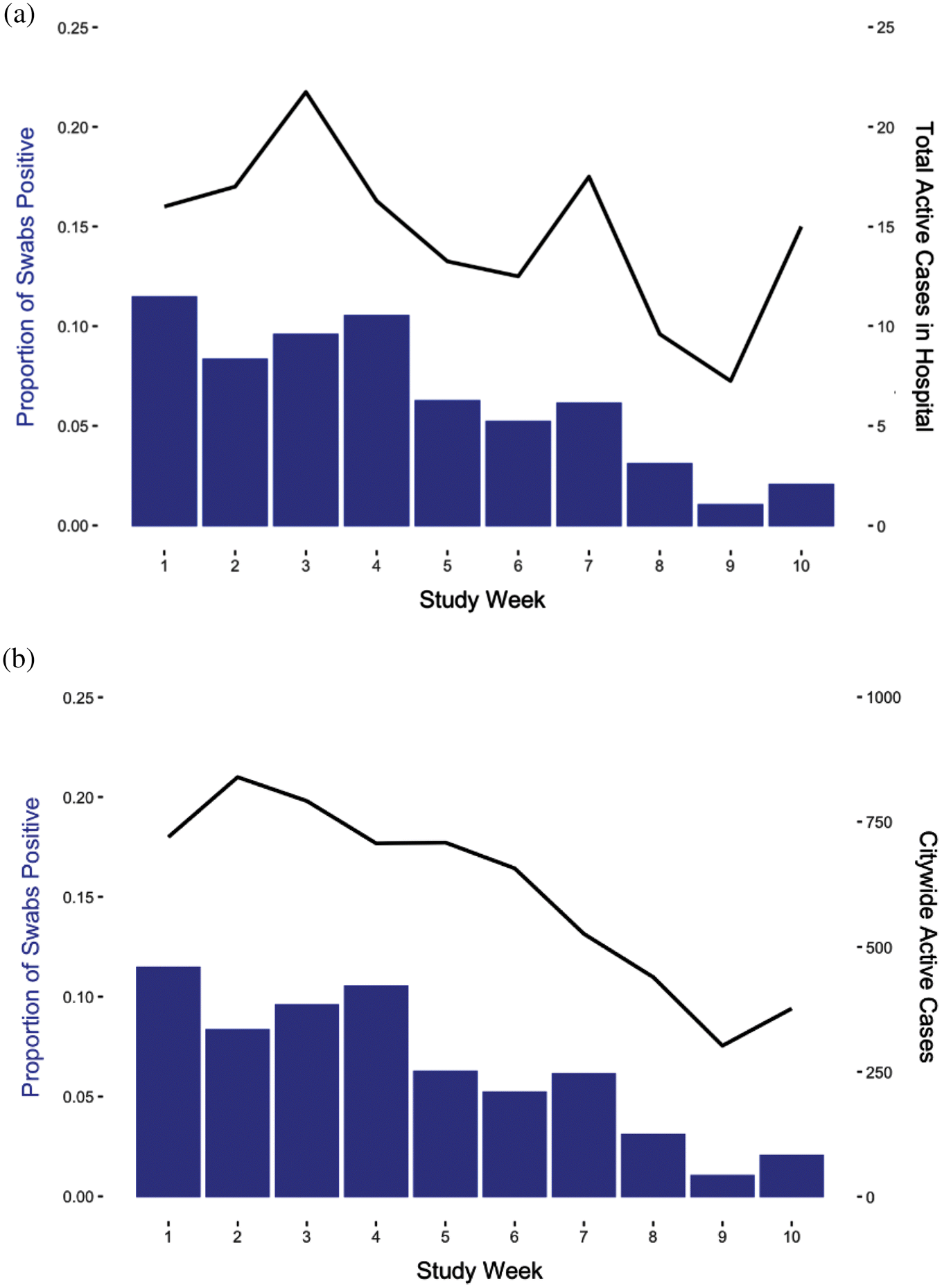
Table 2.
| Characteristic | ORa | 95% CIb | p |
|---|---|---|---|
| Campus | |||
| Campus B | – | – | |
| Campus A | 2.09 | 1.05, 4.15 | 0.04 |
| Unit COVID risk type | |||
| Non-COVID unit | – | – | |
| COVID risk unit | 7.19 | 3.23, 15.99 | <0.01 |
| Public/health care worker area | 1.71 | 0.65, 4.48 | 0.28 |
| Object/site | |||
| Air (Control) | 0 | * | 0.99 |
| Computer keyboard | 0.32 | 0.02, 5.49 | 0.44 |
| Door handle | 0 | * | 0.99 |
| Elevator buttons | 37.56 | 2.95, 479 | <0.01 |
| Exterior bench | 73.11 | 1.93, 2780 | 0.02 |
| Floor | 31.25 | 3.9, 249 | <0.01 |
| Hand sanitizer | 0.48 | 0.03, 8.2 | 0.61 |
| Table | 0 | * | 0.99 |
| Telephone receiver | 0 | * | 0.99 |
| Other | – | – | – |
| Material | |||
| Metal | – | – | – |
| Air (Control) | 0 | * | 0.99 |
| Plastic/vinyl | 8.72 | 1.28, 59.7 | 0.03 |
| Wood | 0.89 | * | 0.99 |
| Study Week | 0.75 | 0.67, 0.85 | <0.01 |
Note
*Inflated variance, CI not reported.
a
OR = Odds Ratio.
b
CI = Confidence interval.
Hospital floors were the most common surface from which SARS-CoV-2 was detected. We looked specifically at prevalence on floors and its relationship with cases within the hospital and citywide (Fig. 4). Restricting analysis to only floors showed they parallel the findings seen with all samples (Fig. 3), with stronger correlation with active hospital cases. Hospital wastewater SARS-CoV-2 viral copies per millilitre (Fig. 5) followed a similar trajectory as built environment sampling, but the decline in wastewater levels appeared to predate the declines in both built environment detection as well as Campus A and citywide case burden.
Fig. 4.
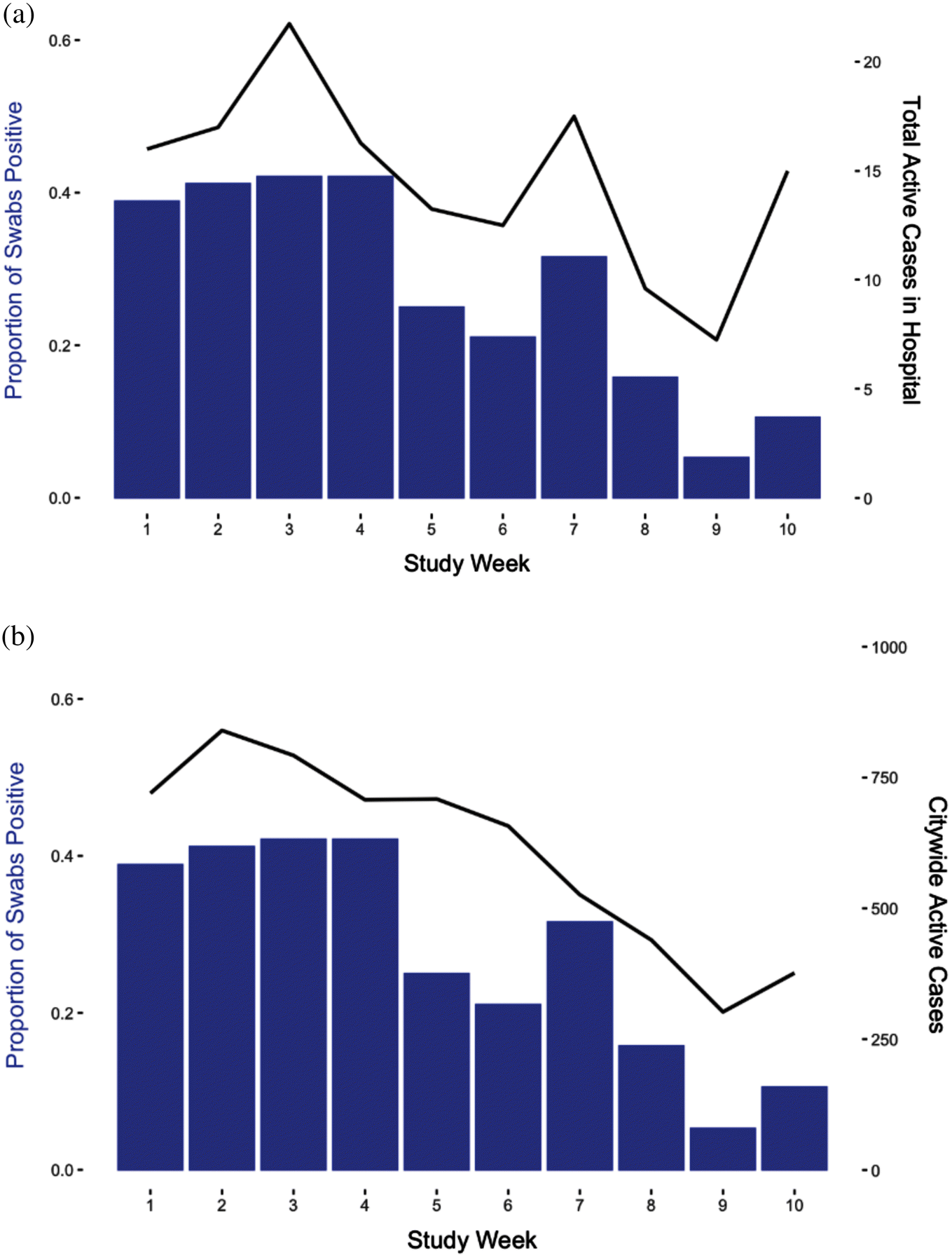
Fig. 5.
We also evaluated the strength of association between (i) all built environment swab positivity or (ii) floor swab positivity with hospital case burden (Supplementary Material 1, Table S2). We found that floor swabs best described the hospital case burden, having a measure of association with the greatest significance and a model with the best fit.
Discussion
In this prospective longitudinal study at two large tertiary care hospital campuses, we demonstrate that SARS-CoV-2 RNA can be readily identified from a variety of hospital surfaces as well as hospital wastewater. Moreover, both the relative frequency of isolation on surfaces and the viral titre in wastewater parallel the burden of infection within the hospital. Wards that specifically managed COVID-19 patients had consistently higher levels of detection compared with other areas, and frequency of detection paralleled case counts over time. For the single ward-based outbreak that occurred during the study period, there was detectable viral RNA at the involved location from the onset and until resolution.
Prior studies have demonstrated the amplification of SARS-CoV-2 RNA from the hospital environment (Guo et al. 2020) including high-touch surfaces outlined in this paper (e.g., elevator buttons, computer keyboards, alcohol hand pumps). Floors have been identified as a common location for identifying SARS-CoV-2 in hospitals and other built environments, and our results reinforce these findings (Guo et al. 2020; D’Accolti et al. 2020; Maestre et al. 2021; Marotz et al. 2021). Moreover, we found that performing surveillance from floors provided the best marker for predicting hospital burden of infection.
To date, fomites have not been identified as a major contributor to transmission of SARS-CoV-2 in the hospital or other settings (Mondelli et al. 2021). While our PCR-based approach for SARS-CoV-2 RNA detection cannot identify whether viral particles present in our samples were viable or not, our results do indicate that identifying any RNA from high-touch surfaces is relatively rare, further supporting these as unlikely pathways for transmission in hospitals. Floors, however, were a common location for SARS-CoV-2 RNA in our study. It is likely they represent a sink for viral particles which originate from the respiratory droplets/aerosol from patients, health workers, or visitors, and it is unlikely that floors represent a reservoir for transmission (Guo et al. 2020).
Interestingly, the non-COVID ward, Ward 3 on Campus A, that experienced the outbreak had frequent detection of SARS-CoV-2 during the outbreak period, with general absence before and after. This suggests that built environment sampling may offer a way to potentially identify occult outbreaks or transmission occurring within patients/visitors/HCWs or for monitoring outbreak status. The outbreak that occurred in the ED at Campus B did have a brief but nonsustained signal, and we believe this is related to the transient nature of patients (and flux of staff) within the ED. Patients are flowing from the ED itself to either discharge or transfer to other sites within the hospital, thus it is unlikely to sustain a patient-based outbreak within that area. The reason for higher detection of SARS-CoV-2 at Campus A is likely driven by more frequent detection at non-COVID risk unit sites. The reason for this is unclear and may relate to both patient and visitor populations.
Where built environment sampling can provide a resolved view of COVID-19 burden in the hospital, hospital wastewater can provide institution-wide surveillance with the possibility for anticipatory (predictive) signals. Qualitatively, wastewater appeared to anticipate burden by one week, and is on the order of magnitude reported in the literature (Randazzo et al. 2020), though given the limited number of time points, we were not able to confirm this statistically. A growing number of studies have shown the potential benefit of wastewater monitoring in both regional as well as facility-based surveillance (Randazzo et al. 2020; Sharif et al. 2021), and our study links this surveillance as complementary to built environment screening. Combined approaches may ultimately be applicable to other respiratory viruses that cause significant institutional outbreaks, including influenza, though further validation is needed.
Our study has some limitations that have not already been noted. Firstly, our study was performed during a period when there was relatively low activity, with regional case counts ranging from approximately two to seven cases per 100 000 population per week, and it is possible that in higher burden settings there could be saturation of ward-based built environment detection that could limit the correlation with burden or outbreaks. Secondly, outbreaks were relatively rare in our hospital, owing in part to the low regional burden, and this limits our ability to evaluate the ability of built environment screening to consistently detect outbreaks. Thirdly, our approaches only detect viral RNA and not viable virus. Lastly, while our approach was capable of detecting SARS-CoV-2 in environmental surface samples, alternative methods such as droplet digital PCR have demonstrated greater sensitivity in low viral load samples and less interference by reaction inhibitors (Falzone et al. 2020). However, our findings benefit from the generalizability of our approaches which use standard PCR techniques and could be easily and quickly performed in most settings worldwide.
In conclusion, we have demonstrated that the detection of SARS-CoV-2 from wastewater and built environment samples, in particular from floors, parallels hospital burden. Built environment sampling may offer a spatially-resolved method for surveillance of hospital COVID-19 cases which can be paired with wastewater monitoring for a comprehensive picture of facility-wide burden of infection. Further prospective studies are needed to evaluate this approach.
Acknowledgements
We would like to thank Dr. Kathryn Suh and Dr. Raphael Saginur for their support and advice during this study. We would also like to thank the City of Ottawa Wastewater Collection Branch, and in particular Benjamin Musyoka and Sandra Gay, for accommodating the collection of wastewater samples. This work was supported by The Ottawa Hospital Academic Medical Organization (TOHAMO), Alliance Grant # 554478 - 20 from the Natural Sciences and Engineering Research Council (NSERC), Carleton University Rapid Response Research Grant, and the Jarislowsky Foundation.
References
Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. 2021. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. New England Journal of Medicine, 384(5): 403–416.
Centers for Disease Control and Prevention. 2020. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes [online]: Available from cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html.
D’Accolti M, Soffritti I, Passaro A, Zuliani G, Antonioli P, Mazzacane S, et al. 2020. SARS-CoV-2 RNA contamination on surfaces of a COVID-19 ward in a hospital of Northern Italy: what risk of transmission? European Review for Medical and Pharmacological Sciences, 24(17): 9202–9207.
Falzone L, Musso N, Gattuso G, Bongiorno D, Palermo CI, Scalia G, et al. 2020. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. International Journal of Molecular Medicine, 46(3): 957–964.
Gonçalves J, Koritnik T, Mioč V, Trkov M, Bolješič M, Berginc N, et al. 2021. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Science of The Total Environment, 755: 143226.
Guo Z-D, Wang Z-Y, Zhang S-F, Li X, Li L, Li C, et al. 2020. Aerosol and surface distribution of Severe Acute Respiratory Syndrome Coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerging Infectious Diseases, 26(7): 1586–1591.
Islam MS, Rahman KM, Sun Y, Qureshi MO, Abdi I, Chughtai AA, et al. 2020. Current knowledge of COVID-19 and infection prevention and control strategies in healthcare settings: A global analysis. Infection Control & Hospital Epidemiology, 41(10): 1196–1206.
Klymus KE, Merkes CM, Allison MJ, Goldberg CS, Helbing CC, Hunter ME, et al. 2020. Reporting the limits of detection and quantification for environmental DNA assays. Environmental DNA, 2(3): 271–282.
Maestre JP, Jarma D, Yu J-RF, Siegel JA, Horner SD, and Kinney KA. 2021. Distribution of SARS-CoV-2 RNA signal in a home with COVID-19 positive occupants. Science of The Total Environment, 778: 146201.
Marotz C, Belda-Ferre P, Ali F, Das P, Huang S, Cantrell K, et al. 2021. SARS-CoV-2 detection status associates with bacterial community composition in patients and the hospital environment. Microbiome, 9(1): 132.
Medema G, Heijnen L, Elsinga G, Italiaander R, and Brouwer A. 2020. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environmental Science & Technology Letters, 7(7): 511–516.
Mondelli MU, Colaneri M, Seminari EM, Baldanti F, and Bruno R. 2021. Low risk of SARS-CoV-2 transmission by fomites in real-life conditions. The Lancet Infectious Diseases, 21(5): e112.
Ontario Ministry of Health. 2020a. COVID-19 reference document for symptoms [online]: Available from health.gov.on.ca/en/pro/programs/publichealth/coronavirus/docs/2019_reference_doc_symptoms.pdf.
Ontario Ministry of Health. 2020b. COVID-19 guidance: Acute care [online]: Available from health.gov.on.ca/en/pro/programs/publichealth/coronavirus/docs/2019_acute_care_guidance.pdf.
Ottawa Public Health. 2021. Daily COVID-19 dashboard [online]: Available from ottawapublichealth.ca/en/reports-research-and-statistics/daily-covid19-dashboard.aspx.
Parker CW, Singh N, Tighe S, Blachowicz A, Wood JM, Seuylemezian A, et al. 2020. End-to-end protocol for the detection of SARS-CoV-2 from built environments. mSystems, 5(5): e00771–20.
Public Health Ontario. 2020. Best practices for prevention, surveillance and infection control management of novel respiratory infections in all health care settings [online]: Available from publichealthontario.ca/-/media/documents/b/2020/bp-novel-respiratory-infections.pdf?la=en.
Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, and Sánchez G. 2020. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Research, 181: 115942.
Sharif S, Ikram A, Khurshid A, Salman M, Mehmood N, Arshad Y, et al. 2021. Detection of SARs-CoV-2 in wastewater using the existing environmental surveillance network: A potential supplementary system for monitoring COVID-19 transmission. PLoS ONE, 16(6): e0249568.
Siemieniuk RA, Bartoszko JJ, Ge L, Zeraatkar D, Izcovich A, Kum E, et al. 2020. Drug treatments for covid-19: living systematic review and network meta-analysis. BMJ, 370: m2980.
Smith DRM, Duval A, Pouwels KB, Guillemot D, Fernandes J, Huynh B-T, et al. 2020. Optimizing COVID-19 surveillance in long-term care facilities: a modelling study. BMC Medicine, 18(1): 386.
van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. 2020. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. The New England Journal of Medicine, 382(16): 1564–1567.
Westhaus S, Weber F-A, Schiwy S, Linnemann V, Brinkmann M, Widera M, et al. 2021. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – Suitability for COVID-19 surveillance and potential transmission risks. The Science of the Total Environment, 751: 141750.
World Health Organization. 2021. WHO Coronavirus (COVID-19) dashboard [online]: Available from covid19.who.int.
Supplementary materials
Supplementary Material 1 (DOCX / 61.8 KB)
- Download
- 61.83 KB
Supplementary Material 2 (XLSX / 22.4 KB)
- Download
- 22.45 KB
Information & Authors
Information
Published In

FACETS
Volume 7 • Number 1 • January 2022
Pages: 82 - 97
Editor: Elena P. Ivanova
History
Received: 8 September 2021
Accepted: 25 November 2021
Version of record online: 20 January 2022
Copyright
© 2022 Hinz et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper and in the Supplementary Material.
Key Words
Sections
Subjects
Authors
Author Contributions
All conceived and designed the study.
AH, LX, RJK, DM, and CN performed the experiments/collected the data.
AH, BO, RJK, DM, and CN analyzed and interpreted the data.
ED contributed resources.
All drafted or revised the manuscript.
Competing Interests
ED works for DNA Genotek that provided sampling swabs in-kind for this study in an unrestricted fashion. DNA Genotek had no control over the findings, interpretations, or conclusions published in this paper. All other authors have no relevant conflicts to disclose.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Aaron Hinz, Lydia Xing, Evgueni Doukhanine, Laura A. Hug, Rees Kassen, Banu Ormeci, Richard J. Kibbee, Alex Wong, Derek MacFadden, and Caroline Nott. 2022. SARS-CoV-2 detection from the built environment and wastewater and its use for hospital surveillance. FACETS.
7(): 82-97. https://doi.org/10.1139/facets-2021-0139
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Detection of Covid-19 Outbreaks Using Built Environment Testing for SARS-CoV-2
2. Quantitative Reverse Transcription PCR Surveillance of SARS-CoV-2 Variants of Concern in Wastewater of Two Counties in Texas, United States
