Peptidases: promising antifungal targets of the human fungal pathogen, Cryptococcus neoformans
Abstract
Cryptococcus neoformans is a globally important fungal pathogen, primarily inflicting disease on immunocompromised individuals. The widespread use of antifungal agents in medicine and agriculture supports the development of antifungal resistance through evolution, and the emergence of new strains with intrinsic resistance drives the need for new therapeutics. For C. neoformans, the production of virulence factors, including extracellular peptidases (e.g., CnMpr-1 and May1) with mechanistic roles in tissue invasion and fungal survival, constitute approximately 2% of the fungal proteome and cover five classes of enzymes. Given their role in fungal virulence, peptidases represent promising targets for anti-virulence discovery in the development of new approaches against C. neoformans. Additionally, intracellular peptidases, which are involved in resistance mechanisms against current treatment options (e.g., azole drugs), as well as capsule biosynthesis and elaboration of virulence factors, present additional opportunities to combat the pathogen. In this review, we highlight key cryptococcal peptidases with defined or predicted roles in fungal virulence and assess sequence alignments against their human homologs. With this information, we define the feasibility of the select peptidases as “druggable” targets for inhibition, representing prospective therapeutic options against the deadly fungus.
1. Introduction
Cryptococcus neoformans is an encapsulated human fungal pathogen responsible for 15% of HIV/AIDS-related deaths worldwide (Sloan and Parris 2014; Rajasingham et al. 2017). The fungus is found ubiquitously within the environment, generally associated with pigeon droppings and various tree species (e.g., eucalyptus), infecting hosts when spores or desiccated fungal cells are inhaled (May et al. 2016). Once inside the lungs, alveolar macrophages phagocytose fungal cells; however, within immunocompromised individuals, fungal cells can escape from the protective barrier of the innate immune system and disseminate throughout the body (Vu et al. 2019). During dissemination, cryptococcal cells can invade the central nervous system following crossing of the blood–brain barrier (BBB) using three proposed mechanisms: (i) transcytosis (i.e., yeast cells transit directly through endothelial cells), (ii) transcellular traversal (i.e., penetration of pathogens between brain microvascular endothelial cells), and (iii) Trojan Horse-like mechanism (i.e., carriage inside an infected macrophage) (Sorrell et al. 2016; Santiago-Tirado et al. 2017; Casadevall et al. 2018). If left untreated or in the presence of antifungal resistant strains, cryptococcosis leads to meningoencephalitis or inflammation of the meninges, the major cause of mortality and morbidity in immunocompromised individuals. Notably, within immunocompetent individuals, the fungus may be cleared upon inhalation; cause bloodstream, pulmonary, or cutaneous infections; or lay dormant and become activated if the immune status of the host changes (Min et al. 2020; Qu et al. 2020).
Treating cryptococcosis varies depending on infection severity and host immune status, with reliance on three distinct classes of antifungals and their mechanisms of action: (i) polyenes (pore formation and leakage of cellular components), (ii) flucytosine (inhibition of DNA and RNA synthesis), and (iii) azoles (ergosterol synthesis inhibition and cell wall integrity impairment) (Perfect 2017; Geddes-McAlister and Shapiro 2019; Bermas and Geddes-McAlister 2020). The widespread use of antifungal agents in medicine and agriculture promotes the development of antifungal resistance through the evolution of resistant strains (Geddes-McAlister and Shapiro 2019). Moreover, the emergence of new fungal strains and species with intrinsic resistance towards common antifungals presents a growing threat. In general, the degree of resistance developed by fungal pathogens varies depending on the class of antifungals administered, and a primary challenge for the development of new antifungals is target similarity between eukaryotes (i.e., fungus and human).
To overcome these challenges, novel therapeutic strategies are needed. One strategy involves the targeting of C. neoformans virulence factors (e.g., growth temperature, polysaccharide capsule, melanin and extracellular enzymes) as potential anti-virulence targets (Siscar-Lewin et al. 2019; Vu et al. 2019; Aaron et al. 2020). This approach has several advantages, including that virulence factors are well-characterized and disarming these factors, instead of killing the pathogen itself, can reduce rates of antifungal resistance by imposing a weaker selective pressure. Various studies have assessed the possibility of targeting extracellular enzymes to reduce or inhibit virulence of Cryptococcus spp. (Olszewski et al. 2004; Shi et al. 2010; Vu et al. 2014). Likewise, C. neoformans produces an array of extracellular and intracellular peptidases associated with virulence and drug resistance, and exploration of the relevance behind targeting such proteins using inhibitors, conveys promising results (Muller and Sethi 1972; Brueske 1986; Bien et al. 2009; Kryštůfek et al. 2021). However, validation of peptidases as therapeutic targets is difficult because of a limited understanding of precise mechanisms of action, kinetic characteristics, biological roles, and homology with human enzymes. In this review, we present a distribution of peptidases within C. neoformans and describe representative enzymes with relevance in virulence, host invasion, and drug resistance. Moreover, through protein alignment strategies, we identify human homologs of the fungal peptidases and highlight commonalities and differences between the sequences, including conservation of active sites. With this information, we propose promising new protease inhibitor targets that may be leveraged as alternative therapeutic options in the future.
2. Peptidases: roles in fungal virulence and antifungal resistance
Peptidases account for almost 6% of total proteins in the human genome and constitute 1–5% of the genomes for bacteria, viruses, and fungi (Barrett 2004). These enzymes are used as virulence factors by many pathogenic microorganisms, including C. neoformans, and they constitute promising targets to combat antifungal resistance, as they impose a weaker selective pressure, restricting the evolution of resistance. For C. neoformans, the proteome consists of 7,429 proteins (Uniprot database 31 August 2021; uniprot.org) with 158 peptidases reported, representing approximately 2% of the total proteome (UP000010091) (Fig. 1).
Fig. 1.
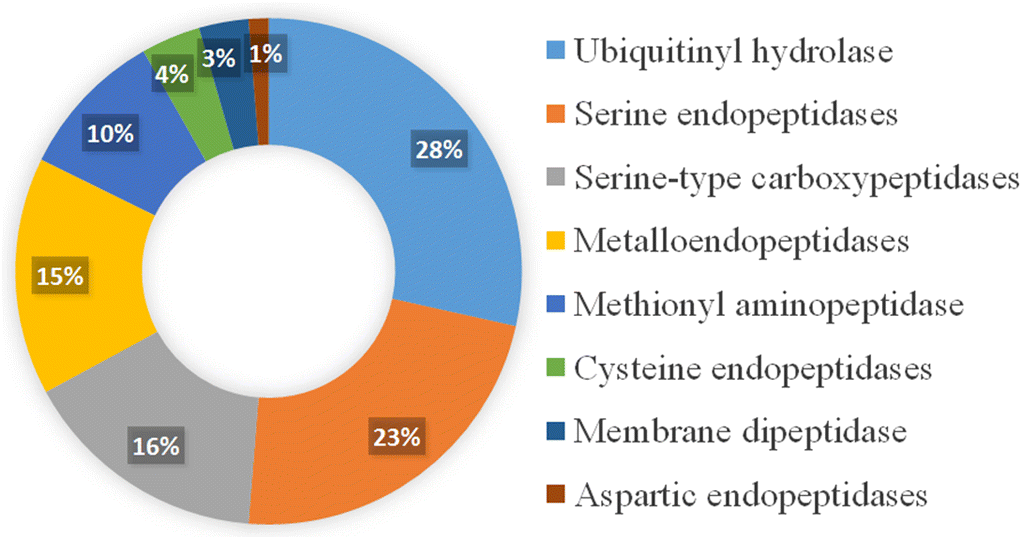
Cryptococcal peptidases compromise several classes, including: (i) endopeptidase, which cleaves peptide bonds within a polypeptide or protein (43%); (ii) ubiquitynil hydrolase, members of the ubiquitin-proteasome pathway (28%); (iii) carboxypeptidase, which cleaves a peptide bond at the carboxy-(C)-terminal of a protein or peptide (16%); (iv) aminopeptidase, which cleaves amino acids from the amino-(N)-terminus of proteins or peptides (10%); and (v) dipeptidase, which cleaves bound pairs of amino acids of proteins or peptides (3%). These classifications support the diverse roles and importance of peptidases within the human fungal pathogen. Specifically, for C. neoformans, extracellular peptidases from different classes assist with a variety of pathogenic roles, from host protein degradation to tissue invasion, and intracellular or membrane-associated peptidases are involved in antifungal resistance and quorum sensing to regulate virulence (Table 1).
Table 1.
| Peptidase | MW (kDa) | Family | Cellular localization | Pathogenic role | Reference |
|---|---|---|---|---|---|
| Carboxypeptidase D (CNAG_00919) | 62 | Serine peptidase (EC: 3.4.16.–) | Extracellular | Host protein degradation, melanin synthesisa | Steen et al. (2002); Clarke et al. (2016) |
| Cerevisin (CNAG_04625) | 53.6 | S8B subtilisin-like serine peptidase (EC: 3.4.21.–) | Extracellular | Melanin synthesis | Eigenheer et al. (2007); Clarke et al. (2016) |
| May1 (CNAG_05872) | 51.3 | A1 Pepsin-like aspartyl peptidase (EC:3.4.23.–) | Extracellular | Immune system evasion, brain invasion, biofilm formationa | Clarke et al. (2016) |
| Pqp1 (CNAG_00150) | 100.9 | S8 subtilisin-like (EC: 3.4.21.–) | Membrane-associated | Quorum sensing | Homer et al. (2016) |
| Rim13 (CNAG_05601) | 89.6 | C2A cysteine peptidase (EC: 3.4.22.–) | Intracellular | Capsule synthesis | Caza and Kronstad (2019) |
| CnMpr-1 (CNAG_04735) | 70 | M36 metallo-peptidase (EC: 3.4.24.–) | Extracellular | Brain invasion | Vu et al. (2014) |
| Stp1 (CNAG_05742) | 65.1 | Metallo-peptidase (EC: 3.4.24.85) | Intracellular | Antifungal resistance | Bien et al. (2009) |
a
Putative.
2.1. Melanin production, thermotolerance, and oxidative damage protection
Melanin is a heterogeneous pigment formed by the oxidative polymerization of indolic or phenolic compounds, protecting cryptococcal cells against oxygen- and nitrogen-derived oxidants produced by host effector cells (Wang and Casadevall 1994). Beyond absorption of free radical fluxes, melanin also contributes to acquired resistance against antifungals, such as Amphotericin B and Caspofungin (Ikeda et al. 2003; van Duin et al. 2020). Furthermore, cryptococcal cells are mesophilic (i.e., grow well at moderate temperatures) and, thus, not well adapted to survive inside the mammalian host where temperature is high (e.g., 37 °C in humans) (Maliehe et al. 2020). Melanin protects fungal cells from elevated temperatures and permits harvesting of host thermal energy to fulfil fungal energetic needs (Dadachova et al. 2007). In C. neoformans, melanin is produced by oxidation of L-3,4-dihyroxyphenylalanine (L-DOPA) via laccase, initiating a series of spontaneous reactions that polymerize the pigment (Eisenman and Casadevall 2012).
2.1.1. Serine-type carboxypeptidase D
During melanin formation, an elevation of enzymes (e.g., hydroxylases or laccases) is observed, as well as an increase in peptidase activity, like carboxypeptidase D (CNAG_00919) and subtilisin-like serine peptidases (Clarke et al. 2016). Although, precise mechanisms of peptidase involvement in melanin production are unclear, subtilisin-like serine peptidases may impact the maturation of enzymes related with phenolic metabolism. For instance, given the role of subtilisin members from higher eukaryotes (e.g., mammals or yeasts) as proprotein convertases (e.g., kexin family or S8B subfamily), a family of proteins that activate other proteins, this may support a connection to melanin production (Gongora et al. 2021). Conversely, carboxypeptidase D, which possesses broad specificity and is responsible for exopeptidase activity outside the cell, may provide free amino acids, such as tyrosine (precursor of L-DOPA), from extracellular protein sources and contribute to melanin synthesis (Clarke et al. 2016).
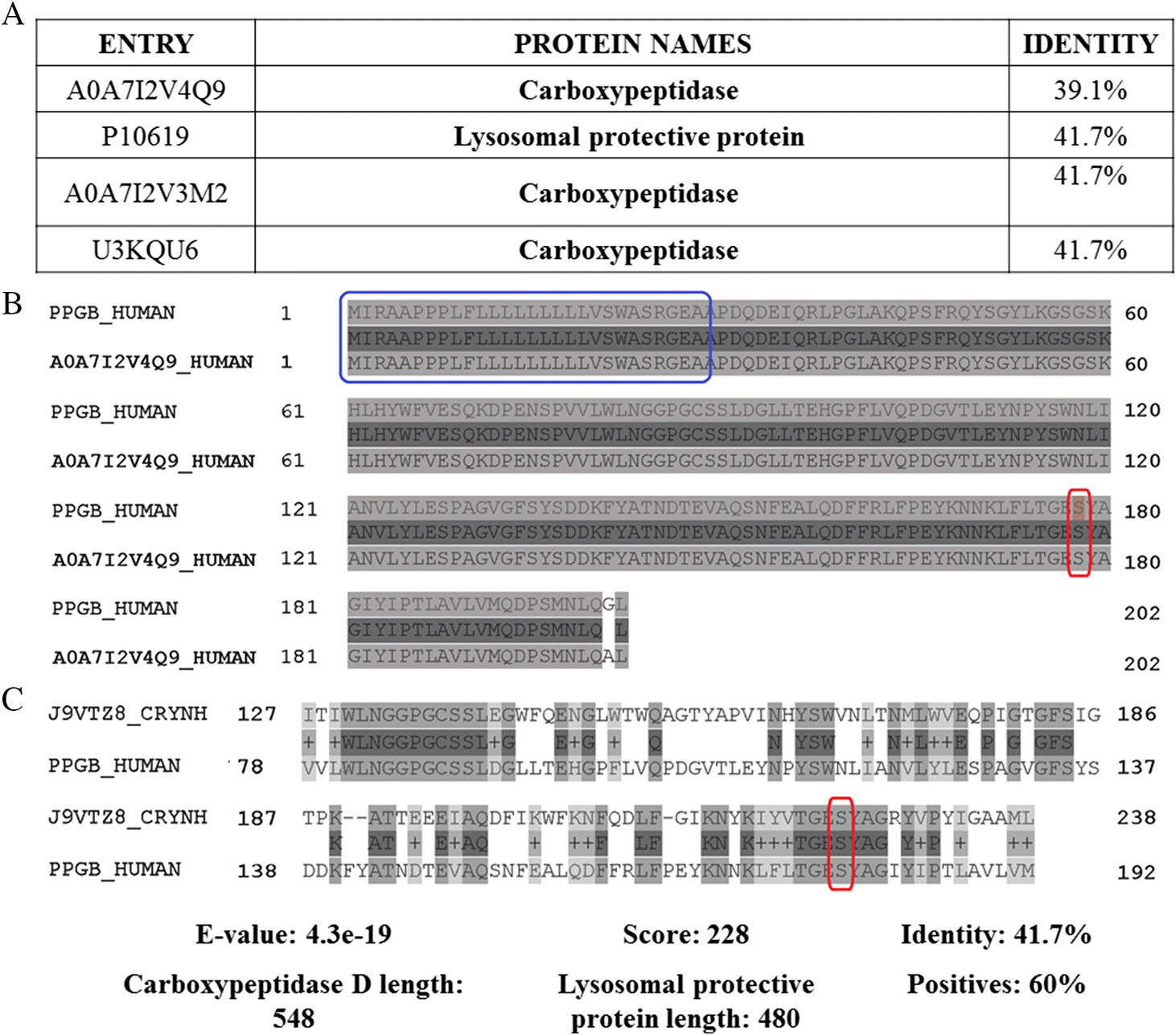
Carboxypeptidase D (EC: 3.4.16.–) is a serine peptidase belonging to the S10 family (Rawlings et al. 2018). Although a homology search indicated that human carboxypeptidase is closely related to the fungal enzyme, the human carboxypeptidase shares 99.5% sequence identity (including the signal peptide) with the heavy polypeptide chain of human lysosomal protective protein (EC:3.4.16.5). This information indicates that carboxypeptidase is an isoform of the polypeptide chain (Fig. 2). Considering these results, we selected human lysosomal protective protein (a heterodimer lysosomal carboxypeptidase related to the regulation of stability and activity of ß-galactosidase and neuraminidase) as the closest human protein to fungal carboxypeptidase D for further analysis (Galjart et al. 1991). These enzymes share high sequence identity (41.7%); however, overlapping regions contain only one amino acid from the active center of the human peptidase (S178), suggesting that these enzymes belong to the same family but are not homologs (Fig. 2). In fact, human lysosomal protective protein is also known as carboxypeptidase C, carboxypeptidase L, and cathepsin A (CTSA). These results indicate that the fungal and host proteins share higher sequence similarity on exosites than at the active sites, possessing different catalytic features and specificities, supporting the design of competitive inhibitors. Additionally, both enzymes are situated in different cellular locations (e.g., endoplasmic reticulum, lumen, or membrane for the human enzyme vs. extracellular space for the fungal enzyme), which highlights carboxypeptidase D as a feasible target against cryptococcosis.
Fig. 2.
2.1.2. Kexin-like cerevisin
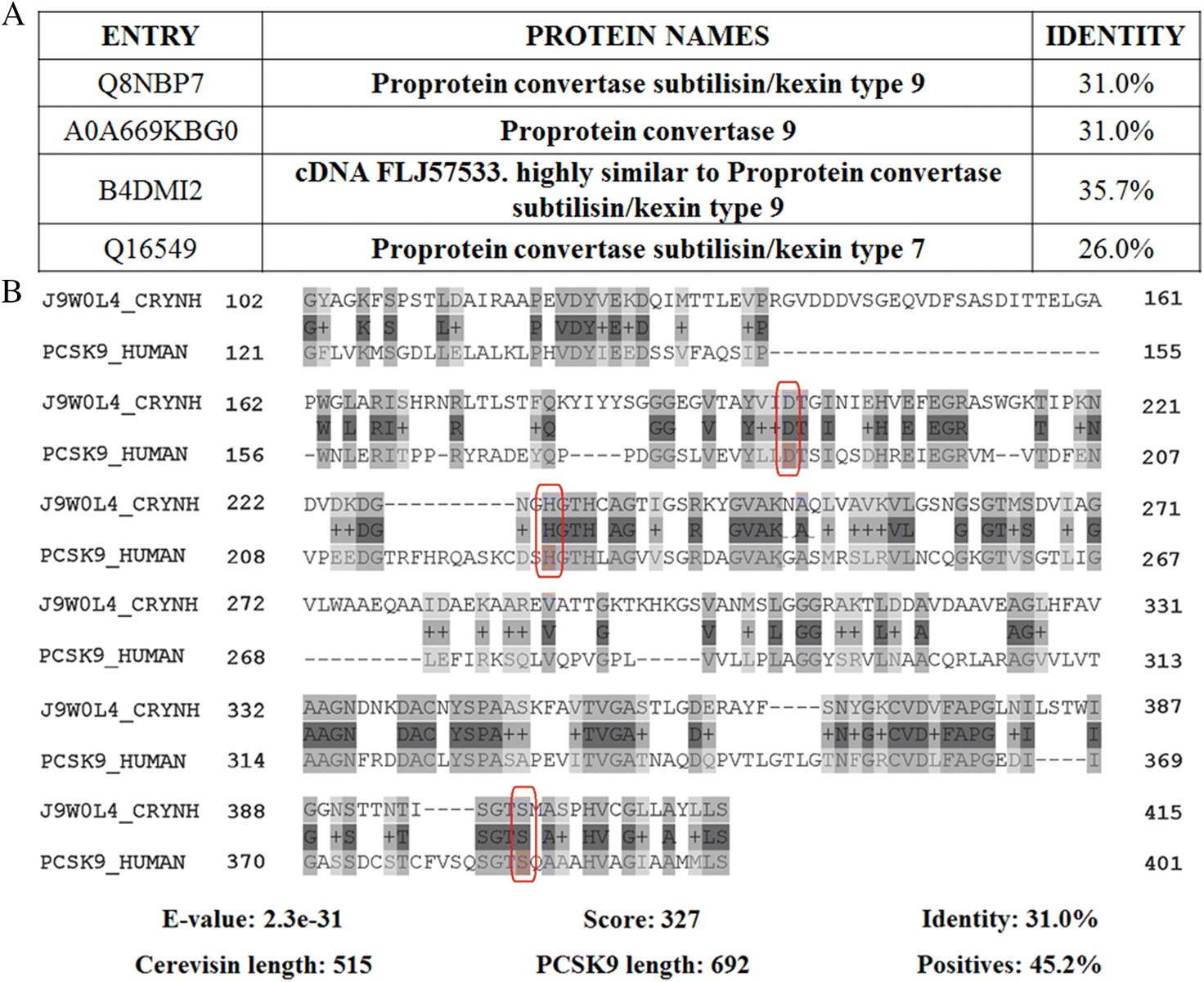
Another candidate peptidase important in melanin production includes cerevisin (CNAG_04625), a secreted serine peptidase identified in the cryptococcal secretome but at baseline levels, suggesting strict substrate specificity and regulation (Clarke et al. 2016). Although a connection with melanin production exists, exact mechanisms of peptidase function and contributions to pigment formation are unknown. Cerevisin (EC: 3.4.21.) is a subtilisin-like serine peptidase belonging to the kexin family (S8B subfamily). Sequence identity with a human homolog, proprotein convertase subtilisin kexin type-9 (PCSK-9) (EC: 3.4.21.61), is 31.7%. PCSK-9 is involved in plasma cholesterol homeostasis regulation by proteolytic degradation of intracellular acidic compartments of lipoprotein and apolipoprotein receptors (e.g., LDLR, LRP1/APOER or LRP8/APOER2) (Fig. 3) (Poirier et al. 2008). The alignment parameters between cerevisin and PCSK-9 are high (e.g., E-value and score) and all amino acids from the active center are conserved, indicating homology between the proteins (Fig. 3) (Pearson 2013). Although this suggests that more research is needed to identify opportunities for leverage of cerevisin as a novel target for inhibition to treat cryptococcal infections, compounds interacting on exosites (e.g., noncompetitive inhibitors) could be also explored.
Fig. 3.
2.2. Biofilm formation
Biofilm structures offer many microorganisms, including C. neoformans, resistance to the host immune response, environmental stresses, and antimicrobial therapy, as well as serving a role in chronic infections (Martinez and Casadevall 2006, 2007). Additionally, cryptococcal cells form biofilms on medical devices, including ventricle–arterial shunt catheters, peritoneal dialysis fistula, cardiac valves, and prosthetic joints, within a clinical setting, posing a substantial threat to patient health (Braun et al. 1994; Banerjee et al. 1997; Johannsson and Callaghan 2009). Notably, after biofilm formation, C. neoformans secretes peptidases (e.g., methionine aminopeptidase and elastinolytic metallopeptidase) involved in fungal invasion, degrading host protein components of the basement membrane (e.g., fibronectin) and extracellular matrix (Santi et al. 2014).
2.2.1. Pepsin-like May1
During biofilm formation, C. neoformans alters protein production to modulate metabolic activity, dormancy, and stress response (Santi et al. 2014). Although peptidases are present at high levels in a biofilm state, a direct relationship between these enzymes and biofilm production has not been well established (Santi et al. 2014). However, previous reports demonstrate that other fungal pathogens (e.g., Candida albicans) secrete aspartic peptidases (e.g., Sap5 and Sap6) at specific stages of biofilm formation to mediate adhesion of fungal cells to surfaces and one another (Kumar et al. 2015; Winter et al. 2016). Aspartic peptidases can also contribute to the breakdown of processing of molecules (e.g., adhesion molecules) important for biofilm formation, such as described in the bacterial pathogen Staphylococcus spp. (Koziel and Potempa 2013; Paharik et al. 2017; Martínez-García et al. 2018). Furthermore, some HIV aspartic peptidase inhibitors constitute effective drugs at preventing biofilm formation and (or) at disrupting established biofilms in C. albicans when combined with major antifungals used to treat infections in the clinic fungi (Lohse et al. 2020). Such observations highlight the importance of serine aspartic peptidases during biofilm formation in fungi.
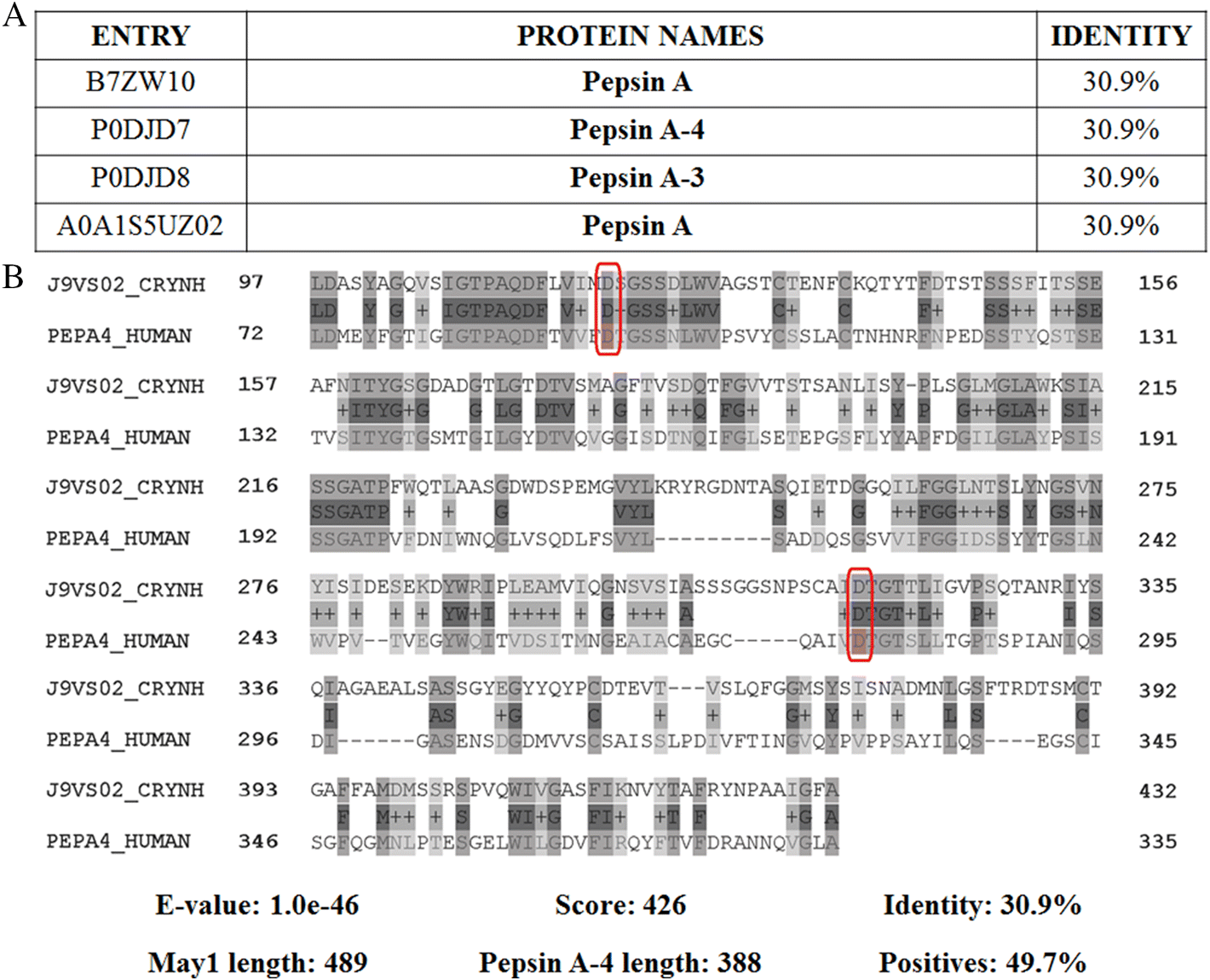
C. neoformans secretes several peptidases, including major aspartyl peptidase 1 (May1), involved in virulence, and possibly degradation of host proteins (Clarke et al. 2016). May1 (CNAG_05872) is an extracellular aspartyl peptidase belonging to the A1 family (Rawlings et al. 2018). Protein sequence alignment of May1 identifies the human protein, Pepsin A-4, sharing sequence identity at 30.9% (Fig. 4). Although these peptidases share the same catalytic amino acids, they share less than 80% of their sequence length, indicating no homology between the proteins (Pearson 2013). These results suggest that May1 could be an ideal antifungal target given the lack of a human homolog; however, both enzymes share active center loop and catalytic amino acids, suggesting similar catalytic properties. Taken together, this information supports the use of noncompetitive inhibitors as potential antifungal compounds, to avoid undesired off-target effects.
Fig. 4.
2.3. Quorum sensing
Quorum sensing is traditionally defined as a bacterial communication mechanism in which secreted signaling molecules impact population function and gene expression (Bassler and Miller 2013). This phenomenon is associated with virulence in some fungi, and in C. neoformans a quorum sensing-like peptide 1 (Qsp1) regulates virulence and cell wall function at high cell densities (Homer et al. 2016). In addition, Qsp1 promotes melanin synthesis, macrophage phagolysosome accumulation, and modulates secreted peptidase activity (e.g., serine-, aspartyl-, and metallo-peptidase) (Homer et al. 2016). This role connects back to the example of May1, an aspartyl peptidase with defined roles in biofilm formation (Clarke et al. 2016), an important example of high cell density structure. Notably, Qsp1 also decreases serine and metallopeptidase activity levels, confirming roles beyond virulence and cell wall function, such as tissue invasion that will be further described below (Homer et al. 2016). Qsp1 is expressed as a pro-peptide, activated extracellularly by peptidase, Pqp1 (CNAG_00150), and internalized, where it exerts its functions as a target of several transcription factors implicated in virulence (Homer et al. 2016).
2.3.1. Subtilisin-like Pqp1
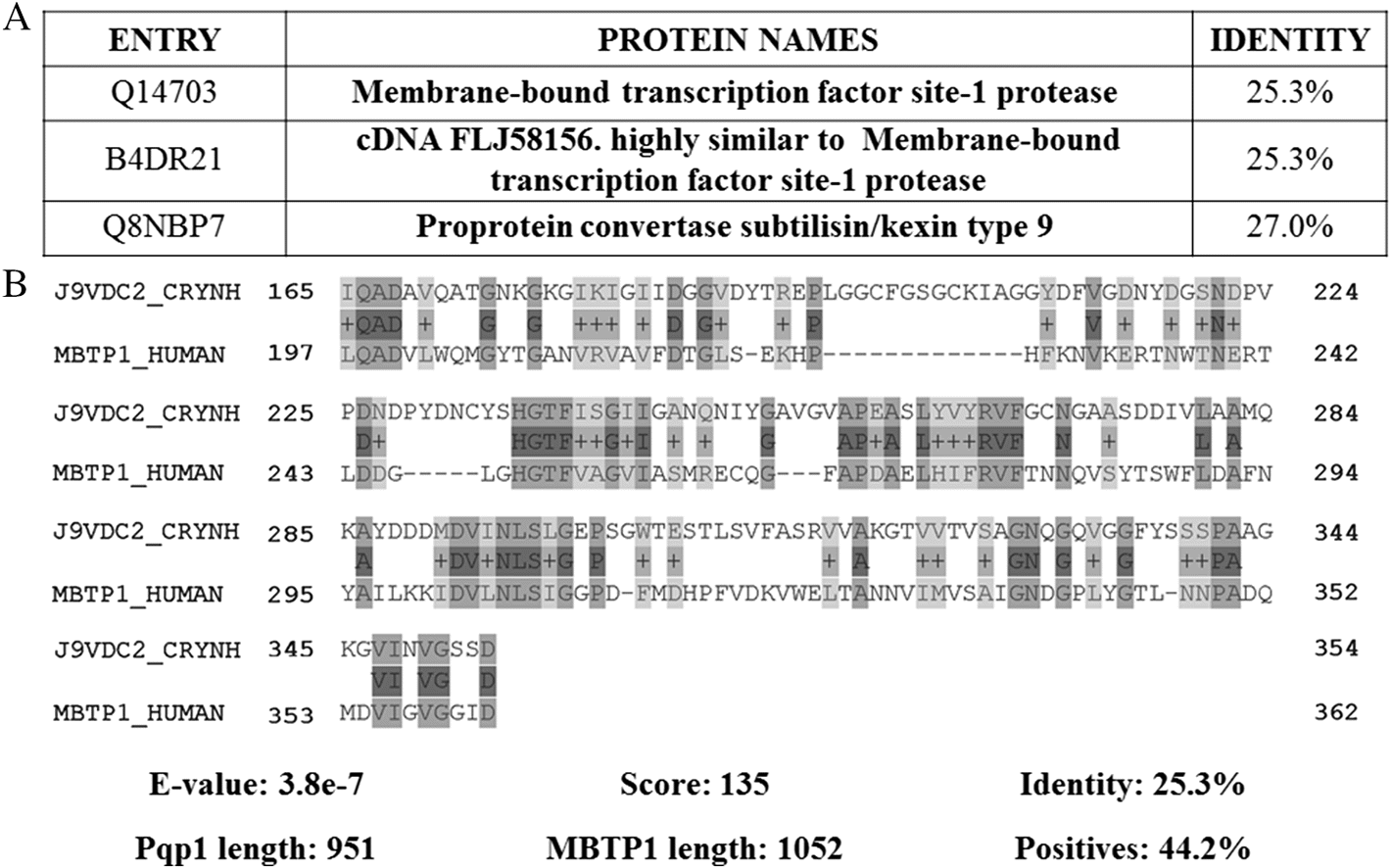
Pqp1 (EC: 3.4.21.–) is a cell-associated peptidase belonging to the subtilisin-like serine peptidase family (S8 family) (Rawlings et al. 2018). This peptidase possesses a low sequence identity with human proteins, with highest identity mapping to a membrane-bound transcription factor site-1 peptidase (MBTP1) (25.3%) (Fig. 5). MBTP1 is an intracellular furin-like peptidase (S8B subfamily) involved in cholesterol metabolism and located in cis- and medial-Golgi (Jaaks and Bernasconi 2017). Considering the relevance of Pqp1 in quorum sensing-like phenomenon to regulate virulence and extracellular peptidase activity (Homer et al. 2016), and its poor similarity with human proteins, Pqp1 arises as an enzyme of interest. Specifically, Pqp1 may serve not only as an anti-cryptococcal target but also to increase knowledge about the quorum sensing process in fungi.
Fig. 5.
2.4. Polysaccharide capsule synthesis
The polysaccharide capsule is an important virulence factor for C. neoformans, contributing to its immunomodulatory and antiphagocytic properties during infection (Bose et al. 2003). The structure is comprised of polysaccharides synthesized intracellularly, exported to the cell surface, and attached to the cell wall (Yoneda and Doering 2006). In C. neoformans, the cyclic-AMP/protein kinase A (cAMP/PKA) signal transduction pathway is a key regulator of capsule elaboration (D’Souza et al. 2001; Kronstad et al. 2011; Caza and Kronstad 2019). Recently, we demonstrated a connection between PKA regulation and the ubiquitin-proteasome pathway (UPP), along with changes in abundance for ribosomal and translational proteins (Geddes et al. 2016). Interestingly, inhibition of the proteasome with Bortezomib affected fungal growth and virulence by impairing capsule formation (Geddes et al. 2016). These findings highlight the role of the proteolytic complex as a potential anti-cryptococcal target and warrant further investigation.
2.4.1. Cysteine-like Rim13
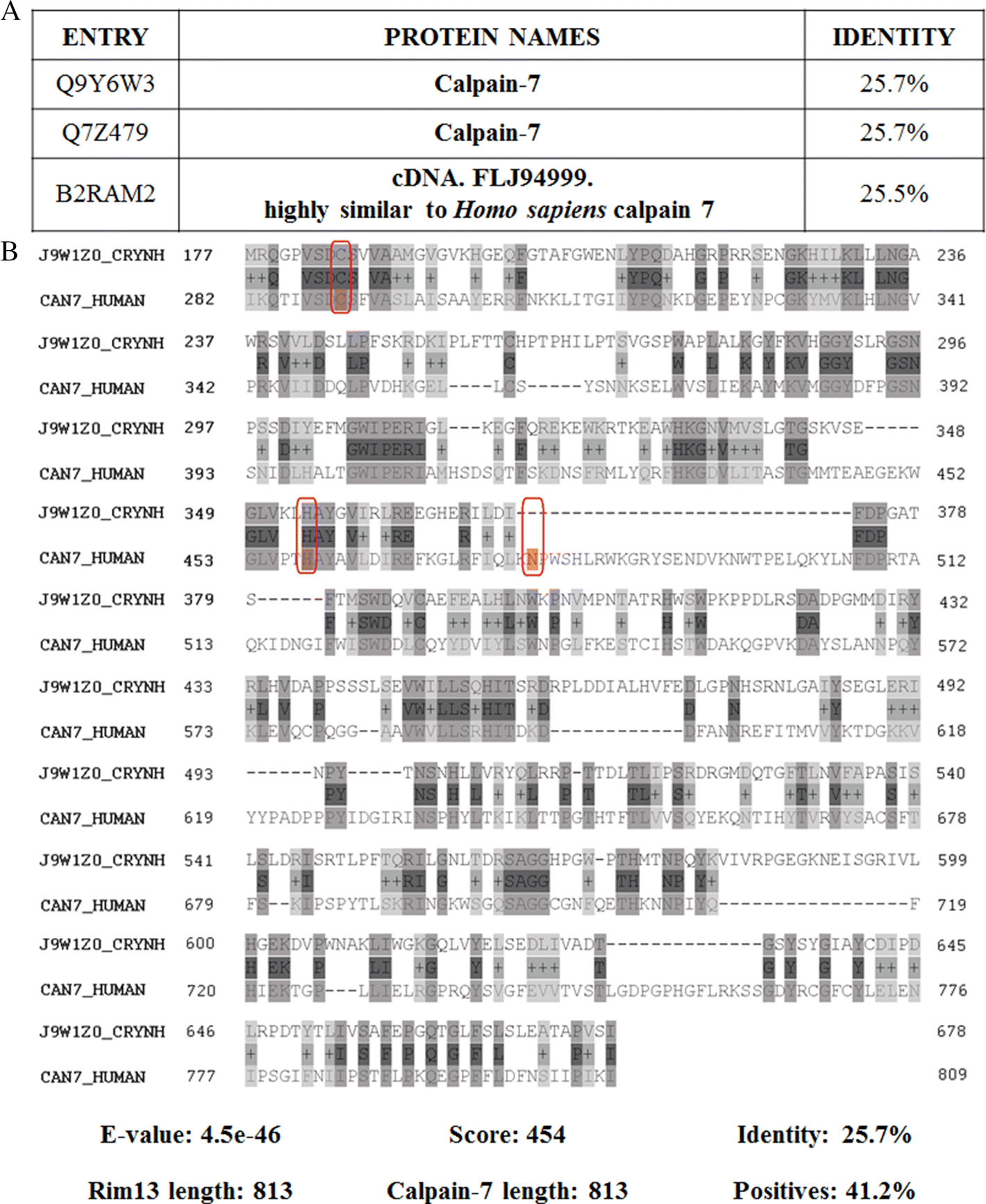
Another important component involved in capsule production includes Rim101, a transcriptional regulator in C. neoformans involved in capsule synthesis, as well as sensing neutral and alkaline extracellular pH as part of the RIM pathway (O’Meara et al. 2010; Ost et al. 2015). This protein requires phosphorylation by PKA before its proteolytic activation by the Rim101 proteolysis complex, which includes Rim13 peptidase (CNAG_05601) (Caza and Kronstad 2019). Once Rim101 is cleaved, it translocates to the nucleus and regulates expression of genes encoding functions for cell wall integrity, metal homeostasis, and capsule attachment to the cell wall (Ost et al. 2015). Rim13 (EC: 3.4.22.–) is a cysteine peptidase belonging to the C2A subfamily, and although it possesses low sequence identity (25.7%) with human proteins, it has the same amino acid length and similar function of Calpain-7 (Fig. 6) (Rawlings et al. 2018). Calpain-7 is a calcium-regulated nonlysosomal thiol-protease involved in the regulation of epithelial cell migration (H. Liu et al. 2013). Importantly, the catalytic amino acids are slightly different, indicating that some enzymatic characteristics, such as binding affinities or specificity, differ between the fungal and human proteins. Such properties highlight an opportunity for selective competitive inhibitors as potential treatment options to avoid off-targets effects.
Fig. 6.
2.5. Central nervous system invasion
C. neoformans possesses specific mechanisms to allow evasion of the human immune system and, in consequence, persist in tissues and organs for prolonged periods in latency (Vu et al. 2019). Dissemination from the lungs to the brain requires migration of fungal cells and crossing of the BBB. This leads to disruption of brain homeostasis and can cause meningoencephalitis, the major pathogenic factor of this fungus (Zaragoza 2019). The mechanism of fungal cell migration, transcytosis, transits fungi directly through endothelial cells, binding to the luminal side of the BBB, facilitating endocytosis (Chen et al. 2003; Shi et al. 2012). This binding and invasion depends on host elements (e.g., CD44 and AnnexinA2), and cryptococcal factors, such as urease, phospholipase, secretion of hyaluronic acid, and peptidases (Santangelo et al. 2004; T.-B. Liu et al. 2013; Vu et al. 2013). For example, serine peptidases have been reported in BBB invasion, degrading human fibronectin, and promoting fungal invasion (Rodrigues et al. 2003; Xu et al. 2014). Moreover, connections between virulence factors, including biofilms and capsule synthesis, can be influenced by peptidases. This is observed for May1, which is active within the acidic environment of macrophages, aiding in fungal survival by degrading host proteins (Chen et al. 1996; Levitz et al. 1999; Clarke et al. 2016). These properties promote immune system evasion, and subsequent brain invasion, confirming the importance of this enzyme as a promising anti-cryptococcal target.
2.5.1. Metallopeptidase, CnMpr-1
CnMpr-1 (CNAG_04735) is a secreted metallopeptidase from C. neoformans required for specific association with BBB and posterior crossing (Vu et al. 2014; Pombejra et al. 2017). This enzyme alters the surface proteins of brain microvascular endothelial cells to promote internalization of fungal cells by inducing host cell surface ruffling (Vu et al. 2014; Pombejra et al. 2017). Additionally, CnMpr-1 engages with cytoskeleton-related proteins, such as AnnexinA2 (AnxA2), inducing transcytosis of cryptococcal cells across endothelial cells cytoplasm, and towards final exocytosis (Pombejra et al. 2017).
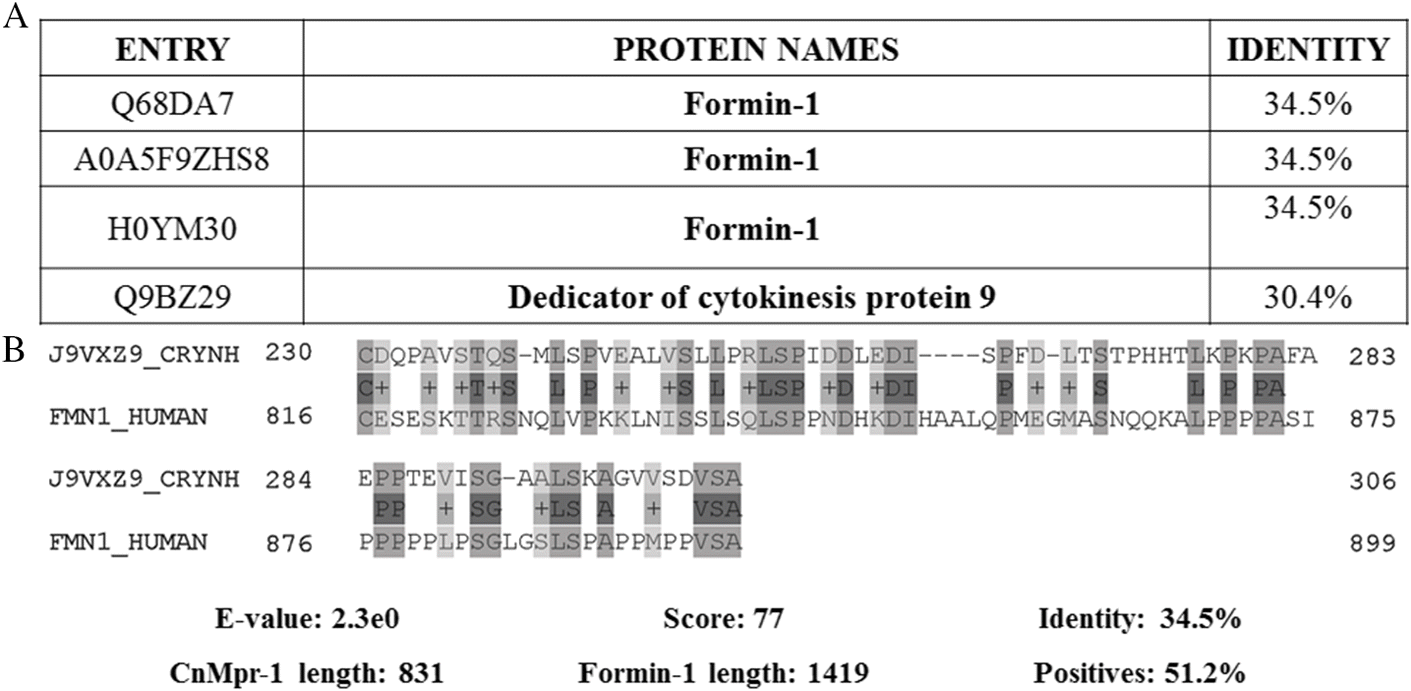
CnMpr-1 belongs to the M36 fungalysins family, which are found only in select fungal species, such as Aspergillus fumigatus and C. neoformans (Eigenheer et al. 2007; Fernández et al. 2013). Although several members of the fungalysins family have been discovered, until present, natural substrates or targets have yet to be identified. In humans, nonpeptidase Formin-1 (involved in microtubule binding) shows sequence identity of 34.5% towards CnMpr-1 with low alignment parameters and diverging sequence length (Boekhorst and Snel 2007; Pearson 2013) (Fig. 7). Considering relevant roles of CnMpr-1 in brain invasion and low sequence identity with a human protein, CnMpr-1 highlights as a promising anti-cryptococcal druggable target.
Fig. 7.
2.6. Antifungal resistance
Ergosterol is an important component in the plasma membrane structure of fungal cells and, consequently, several antifungal therapies such as the azole class of antifungals target the ergosterol synthesis pathway (Gauwerky et al. 2009). Cryptococcal cells lacking ergosterol have a weaker plasma membrane structure and increased cellular permeability, leading to leakage of cellular contents and finally, disruption of fungal growth (Ghannoum and Rice 1999). Although azoles effectively treat cryptococcosis, in recent years, several C. neoformans isolates have developed resistance to these antifungals because of long-term treatment strategies and the fungistatic nature of azoles (i.e., inhibits fungal growth but does not kill the cells) (Smith et al. 2015; Hope et al. 2019; Bermas and Geddes-McAlister 2020).
2.6.1. Metalloprotease, Stp1
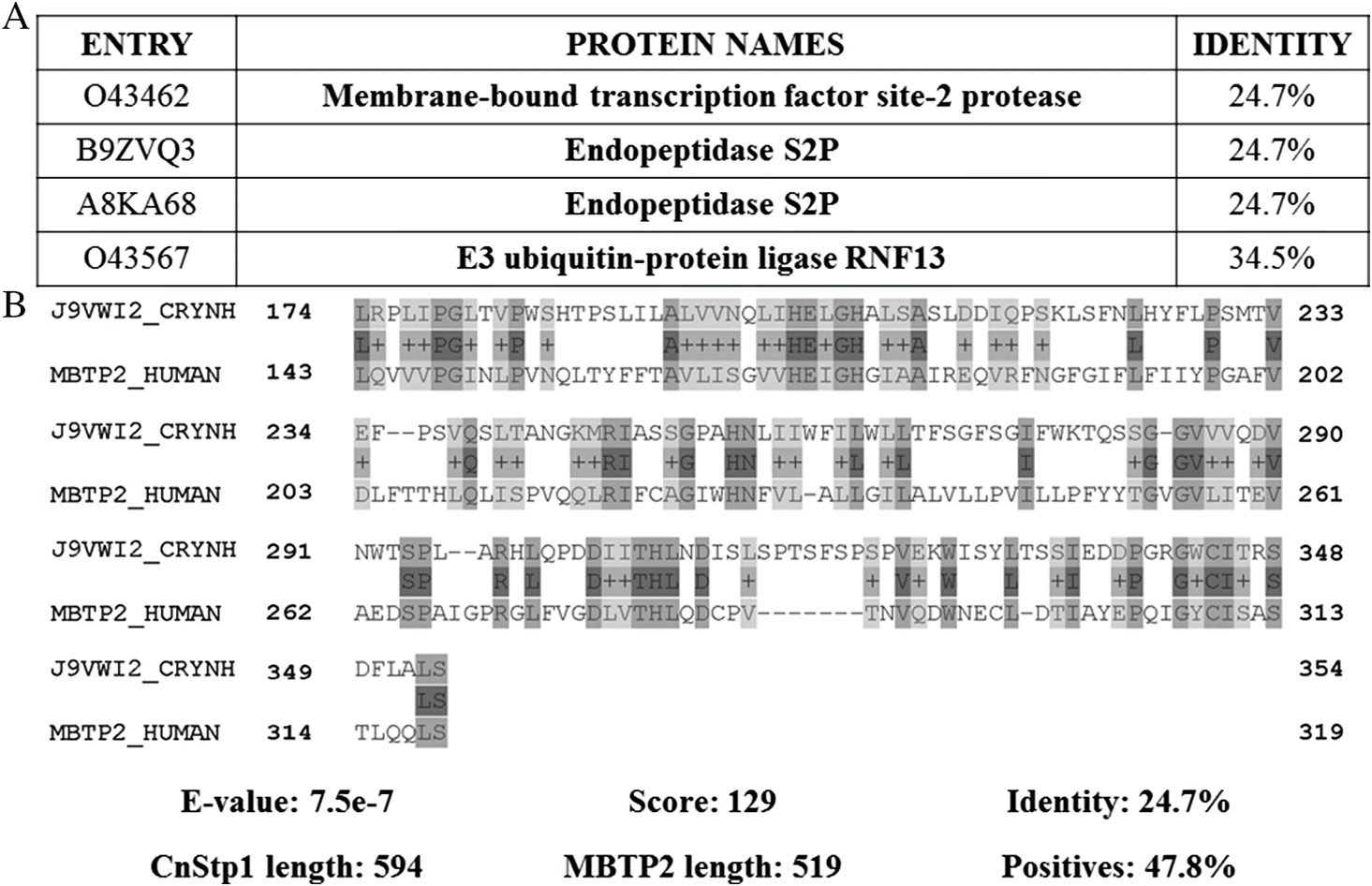
The sterol regulatory element-binding protein (SREBP) pathway is required in C. neoformans for host adaptation and virulence (Chang et al. 2007). Additionally, this factor stimulates ergosterol production in response to sterol depletion, for example when oxygen-dependent ergosterol synthesis is limited by hypoxia (Chun et al. 2007). Furthermore, SREBP factor (Sre1 in C. neoformans) is required for survival in the presence of ergosterol synthesis inhibitors, such as azole class drugs (Chang et al. 2007). Stp1 (CNAG_05742) is a site-2 peptidase relevant in ergosterol synthesis, as is required for Sre1 proteolytic activation, and therefore, antifungal resistance (Bien et al. 2009). Stp1 (EC: 3.4.24.85) is an intracellular metallopeptidase that coordinates the catalytic zinc ion and water molecule in its active site (Feng et al. 2007). Upon protein sequence alignment, the human membrane-bound transcription factor site-2 protease (MBTP2) is identified, sharing a sequence identity of 24.7% with Stp1, suggesting a lack of human homologs (Fig. 8) (Pearson 2013). Given the threat of antifungal resistance in treating infections, highlighting the potential to develop disruptive molecules for Stp1, specific to C. neoformans, suggests a new approach for combatting resistance.
Fig. 8.
3. Cryptococcal peptidases as druggable targets
Peptidases constitute relevant enzymes in several virulent processes in C. neoformans and are also involved in drug resistance mechanisms (See Table 1). The roles and importance of peptidases for fungal pathogenesis have been demonstrated in vitro and using in vivo models of infection (Bien et al. 2009; Vu et al. 2014; Clarke et al. 2016). However, this approach is not applicable in clinical settings; therefore, the design, development, and application of peptidase inhibitors is a practical and feasible alternative. The role of peptidases as potential anti-cryptococcal targets while also highlighting their inhibitors as promising treatment options for further exploration, have been successfully demonstrated in vitro (Aaron et al. 2020; Lohse et al. 2020; Kryštůfek et al. 2021). Moreover, we recently explored the source of such peptidase inhibitors from the natural environment, including plants, invertebrate, and microbes, presenting the strong evolutionary advantage of natural versus synthetic compounds (Gutierrez-Gongora and Geddes-McAlister 2021). Further, we recently presented integration of cryptococcal research at the bench with in vivo models of infection to move applications to the clinic for diagnosis, treatment, and understanding of cryptococcosis (Muselius et al. 2021).
3.1. CnMpr-1 inhibitors prevent BBB crossing
CnMpr-1 (Section 2.5.1), an extracellular metallopeptidase used by cryptococcal cells for specific interactions with the BBB including crossing, invading brain tissue, and causing meningoencephalitis, lacks homology in humans, supporting it as a feasible anti-virulence target for developing therapeutics against cryptococcal meningitis (Vu et al. 2014; Pombejra et al. 2017). In this sense, abietic acid, diosgenin, and lupinine are three natural compounds obtained from plants with inhibitory activity against CnMpr1 (Aaron et al. 2020). In addition, these compounds block BBB crossing by Cryptococcus spp. without affecting endothelial membranes or mammalian cells.
Abietic acid is an abietane diterpenoid found primarily in pine resin, and although its biological activity has not been largely explored, some of its derivatives are known by their antimycotic and antiviral activities (Gonzalez et al. 2010). Other abietane diterpenoids, such as parvifloron D and indigoferabietone, have been described for their anti-cancer and anti-bacterial properties, respectively (Thangadurai et al. 2002; Burmistrova et al. 2015). Diosgenin is a plant steroidal sapogenin used in traditional medicine mainly because of its anti-inflammatory and anti-proliferative activities and anti-cancer properties (Jesus et al. 2016; Sethi et al. 2018). This compound also inhibits the activity of matrix metalloproteinase-2 (MMP-2) and MMP-9, which are involved in the proteolytic processing of matrix proteins (e.g., proteoglycans), adhesion molecules (e.g., integrins), and cell migration (e.g., syndecan) (Koshikawa et al. 2000; Ratnikov et al. 2002; Endo et al. 2003; Li et al. 2004; Chen et al. 2011). Although MMPs have been related with several diseases (e.g., cancer), minimal, if any, cytotoxic effects have been reported against mammalian cells (Bassi et al. 2005; Aaron et al. 2020). These results highlight the importance of further research, mainly focused on increasing specificity, to avoid unwanted off-target effects under normal conditions (e.g., no carcinogen cell presence). Additionally, lupinine, a quinolizidine alkaloid found primarily in the Leguminosae family, displays similar inhibitory properties as abietic acid and diosgenin against CnMpr-1 and shows no presentation of cytotoxic effects (Frick et al. 2017). Other compounds of quinolizidine alkaloid class have been reported as insulin secretion stimulants, which highlights their broad biomedical uses (López et al. 2004).
Importantly, these compounds (i.e., abietic acid, diosgenin, and lupinine) belong to different chemical classes but show similar enzymatic properties against CnMpr-1, suggesting different inhibitory mechanisms. Although it does not possess strong inhibitory activity against CnMpr-1 (half maximal inhibitory concentration (IC50) values around 10 μM) compared with other clinically approved metallopeptidase inhibitors, CnMpr-1 does not currently have effective treatment options (Abbenante and Fairlie 2005). Therefore, these natural compounds constitute a new promising path to target cryptococcal peptidases for inhibition as an alternative strategy and suggest multiple opportunities to achieve the same goal.
3.2. May1 inhibitors could prevent HIV coinfections
Cryptococcosis is the primary fungal-related cause of death among HIV/AIDS patients (Rajasingham et al. 2017). During the HIV replication cycle, proteins require proteolytic processing by HIV-1 peptidase, which belongs to the aspartic protease enzyme class (Dash and Rao 2001). Several anti-HIV treatment options rely on aspartic peptidase inhibitors (e.g., Indinavir, Raquinavir, or Ritonavir) to limit viral replication and reduce viral load and, notably, some also show anti-cryptococcal activity (Blasi et al. 2004; Sidrim et al. 2012). May1 is the major secreted aspartic peptidase of cryptococcal cells and involved in fungal virulence and immune system evasion (Clarke et al. 2016). Therefore, it seems possible that the anti-cryptococcal activity found in HIV-1 peptidase inhibitors may be associated with inhibition of this aspartic peptidase, a positive off-target effect.
Pepstatin A is a classic aspartic peptidase inhibitor and used as a treatment option against fungal-related diseases such as candidiasis and, more recently, as a model to create new derivatives, mainly by inhibition of secreted aspartic peptidases (Braga-Santos and Santos 2011). In this sense, Z-Pst−L-Glu−Hph−NH2 is a pepstatin-derivative peptidase inhibitor that possesses very high inhibitory activity against May1 (Ki = 12 nM) and HIV-1 peptidase (Ki = 32 pM) using similar binding mechanisms (i.e., active site binding) (Kryštůfek et al. 2021). Notably, this compound inhibits both enzymes with similar potency compared with several clinically approved HIV-1 peptidase inhibitors (e.g., Ritonavir or Atazanavir) and presents very high specificity compared with human renin (Ki = 19 μM), its closest structural homologue (Abbenante and Fairlie 2005; Kryštůfek et al. 2021). Although pepstatin A affects C. neoformans and mammalian cell survival with similar potency, suggesting off-target effects, this compound possesses lower cytotoxicity than some clinically applied peptidase inhibitors, including Darunavir, Amprenavir, and Indinavir (Callebaut et al. 2011). These properties support the clinical potential of pepstatin A and warrant further investigation. These results certainly draw attention to Z-Pst−L-Glu−Hph−NH2 as a promising option to develop new treatments against both diseases and (or) to prevent co-infections.
4. Future Directions
In this review, we outlined the distribution of peptidases within the widespread, globally relevant human fungal pathogen, C. neoformans. We selected representative extracellular and intracellular enzymes with described roles in virulence (e.g., melanin and capsule production, biofilm formation, quorum sensing, and dissemination), as well as antifungal resistance. Using protein sequence alignment, we identified human homologs and demonstrated positioning of sequence identity, with an emphasis on catalytic regions, to define potential regions for competitive inhibition. This approach, which combines in vitro and in vivo data with in silico modeling, predictions, and assessment for the rational design of antimicrobial agents is at the forefront of researchers’ minds. The goal is to discover and develop new strategies to overcome antimicrobial resistance and expand our current repertoire of antimicrobial agents (Durand-Reville et al. 2021). Importantly, this strategy brings together interdisciplinary researchers from fields such as microbiology, computer science, biochemistry, molecular biology, chemistry, immunology, and engineering to address the problem of limited therapeutics. However, a large portion of research efforts and funding is still dedicated towards bacterial infections, leaving fungal pathogens with many opportunities to circumvent our current arsenal of defense. Although the need for novel antifungal agents to combat fungal infections of humans, animals, and crops to ensure global health and food security is dire, much can be gleaned from advances made against bacterial pathogens, including the discovery and characterization of novel proteases and effective inhibitors (Fetzer et al. 2017). We also presented examples of peptidase inhibitors used in the clinic with predicted off-target effects against cryptococcal enzymes as well as alternative options with inhibitory activity and improved cytotoxicity effects compared with currently used clinical inhibitors. Together, these peptidase inhibitors may be optimized for fungal-specific activity or as a foundation for additional drug design.
Another critical component in our fight against fungal pathogens and subsequent infections is the acknowledgement that our identification and characterization of peptidases is far from complete. In C. neoformans alone, over 2,000 uncharacterized or hypothetical proteins remain (Uniprot, Fungidb: fungidb.org/fungidb/app), and the potential to discover many more enzymes involved in fungal virulence or antifungal resistance is high, which coincides with an opportunity to develop antifungal agents against these new targets. An important technology for the discovery of novel proteases is mass spectrometry-based proteomics, which detects and quantifies proteins produced under a variety of experimental conditions and has application within both the lab and the clinic (Aebersold and Mann 2016; Zhu et al. 2021). For many fungal pathogens, proteomics is a promising tool to further our understanding of pathogenesis, define signaling networks, explore interactions with the host, develop biomarkers for diagnostics, as well as assess treatment efficacy (Aebersold et al. 2016; Ball et al. 2019; Ball et al. 2020; Muselius et al. 2021; Retanal et al. 2021). Additionally, peptidases produced by the fungus can be measured within the intracellular (cellular proteome) and extracellular (secretome) environments for identification. This can be followed by molecular and biochemical experimentation for characterization, such as genetic manipulation to define a role for the corresponding gene, as well as pull-down or immunoprecipitation assays to identify direct and indirect interaction partners. Moreover, mass spectrometry-based proteomics can profile interactions between a protein and drug or inhibitor, assess binding capacities and detect off-target effects for an important and decisive role in drug discovery (Reinhard et al. 2015). Taken together, the potential for further discovery of fungal peptidases relevant for virulence, as well as the opportunity to design and develop inhibitors for targeted and specific activity is promising.
5. Conclusion
To date, more than 7400 proteins have been reported in C. neoformans proteome where peptidases represent approximately 2% of defined proteins. However, given that approximately 31% of total cryptococcal proteins remain uncharacterized, further research is warranted, and more enzymes, specifically extracellular and intracellular peptidases have yet to be defined and characterized. Cryptococcal peptidases, mainly extracellular, are not only related with other virulence factors like thermotolerance, fungal growth, or melanin synthesis but are also involved in dissemination for brain invasion and immune system evasion. Although intracellular peptidases have been less studied, these enzymes constitute important elements involved in diverse biological processes, such as capsule formation and antifungal resistance. Moreover, some cryptococcal peptidases are implicated in quorum sensing, a phenomenon that is still unclear in fungi but highlights a remarkable process for regulating secreted peptidases and virulence. Overall, these studies show that proteolytic activity is not a random process in C. neoformans pathogenicity but is subject to tight control with many mechanisms remaining to be discovered. Furthermore, based on our analyses described herein, many of these enzymes show low sequence identity with human proteins, underscoring their role as prospective “druggable” anti-cryptococcal targets for further exploration.
Acknowledgments
We thank members of the Geddes-McAlister lab for their critical reading and feedback for this manuscript.
References
Aaron PA, Vu K, and Gelli A. 2020. ‘An antivirulence approach for preventing Cryptococcus neoformans from crossing the Blood-Brain barrier via Novel natural product inhibitors of a fungal metalloprotease’, MBio, 11(4): 1–15.
Abbenante G, and Fairlie DP. 2005. Protease inhibitors in the clinic. Medicinal Chemistry, 1(1): 71–104.
Aebersold R., Bensimon A, Collins BC, Ludwig C, and Sabido E. 2016. Applications and developments in targeted proteomics: from SRM to DIA/SWATH. Proteomics.
Aebersold R, and Mann M. 2016. Mass-spectrometric exploration of proteome structure and function. Nature, 537(7620): 347–355.
Ball B, Bermas A, Carruthers-Lay D, and Geddes-McAlister J, et al. 2019. Mass spectrometry-based proteomics of fungal pathogenesis, host–fungal interactions, and antifungal development. Journal of Fungi, 5(2): 52.
Ball B, Langille M, and Geddes-McAlister J. 2020. Fun(gi)OMICS: advanced and diverse technologies to explore emerging fungal pathogens. MBio, 11(5): e01020–20.
Banerjee U, Gupta K, and Venugopal P. 1997. A case of prosthetic valve endocarditis caused by Cryptococcus neoformans var. neoformans. Journal of Medical and Veterinary Mycology, Informa UK Ltd UK, 35(2): 139–141.
Barrett AJ. 2004. Bioinformatics of proteases in the MEROPS database. Current Opinion in Drug Discovery. Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridgeshire CB10 ISA, UK. 7(3): 334–341.
Bassi DE, Fu J, Lopez de Cicco R, and Klein-Szanto AJP. 2005. Proprotein convertases: master switches in the regulation of tumor growth and progression. Molecular Carcinogenesis, 44: 151–161.
Bassler BL, and Miller MB. 2013. Quorum sensing. In The Prokaryotes: Prokaryotic Communities and Ecophysiology. Springer-Verlag Berlin Heidelberg. Vol. 9783642301230. pp. 495–509.
Bermas A, and Geddes-McAlister J. 2020. Combatting the evolution of anti-fungal resistance in Cryptococcus neoformans. Molecular Microbiology, 114(5): 721–734. Available at: onlinelibrary.wiley.com/doi/full/10.1111/mmi.14565.
Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, and Espenshade PJ, et al. 2009. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Molecular Microbiology, 74: 672–690.
Blasi E, Colombari B, Orsi CF, Pinti M, Troiano L, Cossarizza A, et al. 2004. The human immunodeficiency virus (HIV) protease inhibitor indinavir directly affects the opportunistic fungal pathogen Cryptococcus neoformans. FEMS Immunology and Medical Microbiology, 42: 187–195.
Boekhorst J, and Snel B. 2007. Identification of homologs in insignificant blast hits by exploiting extrinsic gene properties. BMC Bioinformatics, 8(356).
Bose I., Reese AJ, Ory JJ, Janbon G, and Doering TL. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryotic Celltic Cell, 2: 655–663.
Braga-Santos LA, and Santos ALS. 2011. Aspartic protease inhibitors as potential anti- Candida albicans drugs: impacts on fungal biology, virulence and pathogenesis. Current Medicinal Chemistry, 18: 2401–2419.
Braun DK, Janssen DA, Marcus JR, and Kauffman CA. 1994. Cryptococcal infection of a prosthetic dialysis fistula. American Journal of Kidney Diseases, Elsevier, 24(5): 864–867.
Brueske CH. 1986. Proteolytic activity of a clinical isolate of Cryptococcus neoformans. Journal of Clinical Microbiology, 23(3): 631–633.
Burmistrova O, Perdomo J, Simões MF, Rijo P, Quintana J, and Estévez F, et al. 2015. The abietane diterpenoid parvifloron D from Plectranthus ecklonii is a potent apoptotic inducer in human leukemia cells. Phytomedicine, Elsevier, 22(11): 1009–1016.
Callebaut C, Stray K, Tsai L, Williams M, Yang Z-Y, Cannizzaro C, et al. 2011. In vitro characterization of GS-8374, a novel phosphonate-containing inhibitor of HIV-1 protease with a favorable resistance profile. Antimicrobial Agents and Chemotherapy, Am Soc Microbiol, 55(4): 1366–1376.
Casadevall A, Coelho C, and Alanio A. 2018. Mechanisms of Cryptococcus neoformans – mediated host damage. Frontiers in Immunology, 9(April): 1–8.
Caza M, and Kronstad JW. 2019. The cAMP/protein kinase a pathway regulates virulence and adaptation to host conditions in Cryptococcus neoformans. Frontiers in Cellular and Infection Microbiology.
Chang YC, Peek CB, Espenshade PJ, Kwon-Chung KJ, and Lee H, et al. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Molecular Microbiology, 64: 614–629.
Chen L, Blank ES, and Casadevall A. 1996. Extracellular proteinase activity of Cryptococcus neoformans. Clinical and Diagnostic Laboratory Immunology, 3(5): 570–574.
Chen P, Shih Y-W, Huang H-C, and Cheng H-W. 2011. Diosgenin, a steroidal saponin, inhibits migration and invasion of human prostate cancer PC-3 cells by reducing matrix metalloproteinases expression. PLoS ONE, 6: e20164.
Chen SHM, Stins MF, Huang S-H, Chen YH, Kwon-Chung KJ, Chang Y, et al. 2003. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. Journal of Medical Microbiology. Microbiology Society, 52(11): 961–970.
Chun CD, Liu OW, and Madhani HD. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathogens, 3: e22.
Clarke SC, Dumesic PA, Homer CM, O’Donoghue AJ, La Greca F, Pallova L, et al. 2016. Integrated activity and genetic profiling of secreted peptidases in Cryptococcus neoformans reveals an Aspartyl Peptidase required for low pH survival and virulence. PLoS Pathogens.
D’Souza C, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, and Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Molecular Biology Cell, 21: 3179–3191.
Dadachova E, Bryan RA, Huang X, Moadel T, Schweitzer AD, et al. 2007. Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi. PloS One, 2(5): e457.
Dash C, and Rao M. 2001. Interactions of a novel inhibitor from an extremophilic Bacillus sp. with HIV-1 pProtease. The Journal of Biological Chemistry, 276(4): 2487–2493.
Durand-Reville TF, Miller AA, O’Donnell JP, Wu X, Sylvester MA, Guler S, et al. 2021. Rational design of a new antibiotic class for drug-resistant infections. Nature, 597.
Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, and Gelli A. 2007. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Research, 7: 499–510.
Eisenman HC, and Casadevall A. 2012. Synthesis and assembly of fungal melanin. Applied Microbiology and Biotechnology.
Endo K, Takino T, Miyamori H, Kinsen H, Yoshizaki T, Furukawa M, and Sato H, et al. 2003. Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. Journal of Biological Chemistry, ASBMB, 278(42): 40764–40770.
Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, and Shi Y, et al. 2007. Structure of a site-2 peptidase family intramembrane metallopeptidase. Science, 318: 1608–1612.
Fernández D, Russi S, Vendrell J, Monod M, and Pallarès I. 2013. A functional and structural study of the major metallopeptidase secreted by the pathogenic fungus Aspergillus fumigatus. Acta Crystallographica Section D: Biological Crystallography, 69: 1946–1957.
Fetzer C, Korotkov VS, Thänert R, Lee KM, Neuenschwander M, von Kries JP, Medina E, and Sieber SA. 2017. A chemical disruptor of the ClpX Chaperone complex attenuates the virulence of multidrug-resistant Staphylococcus aureus. Angewandte Chemie – International Edition, 56(49): 15746–15750.
Frick K, Kamphuis LG, Siddique KHM, Singh KB, and Foley RC. 2017. Quinolizidine alkaloid biosynthesis in lupins and prospects for grain quality improvement. Frontiers in Plant Science, 8: 87.
Galjart NJ, Morreau H, Willemsen R, Gillemans N, Bonten EJ, and d’Azzo A. 1991. Human lysosomal protective protein has cathepsin A-like activity distinct from its protective function. The Journal of Biological Chemistry, 266: 14754–14762.
Gauwerky K, Borelli C, and Korting HC. 2009. Targeting virulence: a new paradigm for antifungals. Drug Discovery Today. Elsevier, 14(3–4): 214–222.
Geddes-McAlister J, and Shapiro RS. 2019. New pathogens, new tricks: emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Annals of the New York Academy of Sciences, 1435(1): 57–78.
Geddes JMH, Caza M, Croll D, Stoynov N, Foster LJ, and Kronstad JW, et al. 2016. Analysis of the protein kinase a-regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin-proteasome pathway in capsule formation. mBio, 7(1): 1–15.
Ghannoum MA, and Rice LB. 1999. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clinical Microbiology Reviews, 12(4): 501–517.
Gongora DG, Perez LR, Muñoz AC, and del Rivero Antigua MA, et al. 2021. Biomedical and Biological relevance of subtilases from the S8 family, diverse and widely distributed serine peptidases. Revista Cubana de Ciencias Biológicas, 9(2): 1–13.
Gonzalez MA, Pérez-Guaita D, Correa-Royero J, Zapata B, Agudelo L, Mesa-Arango A, and Betancur-Galvis L. 2010. Synthesis and biological evaluation of dehydroabietic acid derivatives. European Journal of Medical Chemistry, 45: 811–816.
Gutierrez-Gongora D, and Geddes-McAlister J. 2021. From naturally-sourced protease inhibitors to new treatments for fungal infections. Journal of Fungi, 7(12).
Homer CMM, Summers DK, Goranov AI, Clarke SC, Wiesner DL, Diedrich JK, Moresco JJ, et al. 2016. Intracellular action of a secreted peptide required for fungal virulence. Cell Host & Microbe, 19(6): 849–864.
Hope W, Stone NRH, Johnson A, McEntee L, Farrington N, Santoro-Castelazo A, Liu X, et al. 2019. Fluconazole monotherapy is a suboptimal option for initial treatment of cryptococcal meningitis because of emergence of resistance. MBio, 10(6): e02575–19.
Ikeda R, Sugita T, Jacobson ES, and Shinoda T. 2003. Effects of melanin upon susceptibility of Cryptococcus to antifungals. Microbiology and Immunology, 47: 271–277.
Jaaks P, and Bernasconi M. 2017. The proprotein convertase furin in tumour progression. International Journal of Cancer, 141: 654–663.
Jesus M, Martins APJ, Gallardo E, and Silvestre S. 2016. Diosgenin: recent highlights on pharmacology and analytical methodology. Journal of Analytical Methods in Chemistry, 2016: 1.
Johannsson B, and Callaghan JJ. 2009. Prosthetic hip infection due to Cryptococcus neoformans: case report. Diagnostic Microbiology and Infectious Disease, 64(1): 76–79.
Koshikawa N, Giannelli G, Cirulli V, Miyazaki K, and Quaranta V. 2000. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. Journal of Cell Biology, 148(3): 615–624.
Koziel J, and Potempa J. 2013. Protease-armed bacteria in the skin. Cell and Tissue Research, 351(2): 325–337.
Kronstad JW, Attarian R, Cadieux B, Choi J, D’Souza CA, Griffiths EJ, et al. 2011. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nature Reviews Microbiology, 9: 193–203.
Kryštůfek R, et al. 2021. Re-emerging Aspartic Protease targets: examining Cryptococcus neoformans major Aspartyl Peptidase 1 as a target for antifungal drug discovery. Journal of Medicinal Chemistry, 64: 6706–6719.
Kumar R, Saraswat D, Tati S, and Edgerton M. 2015. Novel aggregation properties of Candida albicans secreted aspartyl proteinase Sap6 mediate virulence in oral candidiasis. Infection and Immunity, 83(7): 2614–2626.
Levitz SM, Nong S-H, Seetoo KF, Harrison TS, Speizer RA, and Simons ER. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infection and Immunity, 67(2): 885–890.
Li Y, Aoki T, Mori Y, Ahmad M, Miyamori H, Takino T, and Sato H. 2004. Cleavage of lumican by membrane-type matrix metalloproteinase-1 abrogates this proteoglycan-mediated suppression of tumor cell colony formation in soft agar. Cancer Research, 64(19): 7058–7064.
Liu H, Jiang Y, Jin X, Zhu L, Shen X, Zhang Q, et al. 2013. CAPN 7 promotes the migration and invasion of human endometrial stromal cell by regulating matrix metalloproteinase 2 activity. Reproductive Biology and Endocrinology, 11(64): 1–8.
Liu TB, Kim J-C, Wang Y, Toffaletti DL, Eugenin E, Perfect JR, Kim KJ, and Xue C. 2013. Brain inositol is a novel stimulator for promoting Cryptococcus penetration of the blood-brain barrier. PLoS Pathogens, 9(4): e1003247.
Lohse MB, Gulati M, Craik CS, Johnson AD, and Nobile CJ. 2020. Combination of antifungal drugs and protease inhibitors prevent Candida albicans biofilm formation and disrupt mature biofilms. Frontiers in Microbiology, 11(1027): 1–12.
López PMG, de la Mora PG, Wysocka W, Maiztegui B, Alzugaray ME, Del Zotto H, and Borelli MI. 2004. Quinolizidine alkaloids isolated from Lupinus species enhance insulin secretion. European Journal of Pharmacology, 504(1–2): 139–142.
Maliehe M, Ntoi MA, Lahiri S, Folorunso OS, Ogundeji AO, Pohl CH, and Sebolai OM, et al. 2020. Environmental factors that contribute to the maintenance of Cryptococcus neoformans pathogenesis. Microorganisms, 8(180): 1–19.
Martinez LR, and Casadevall A. 2006. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrobial Agents and Chemotherapy, 50(3): 1021–1033.
Martinez LR, and Casadevall A. 2007. Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Applied and Environmental Microbiology, 73(14): 4592–4601.
Martínez-García S, Rodríguez-Martínez S, Cancino-Diaz ME, and Cancino-Diaz JC, et al. 2018. Extracellular proteases of Staphylococcus epidermidis: roles as virulence factors and their participation in biofilm. APMIS, 126: 177–185.
May RC, Stone NRH, Wiesner DL, Bicanic T, and Nielsen K, et al. 2016. Cryptococcus: from environmental saprophyte to global pathogen. Nature Reviews Microbiology, 14(2): 106–117.
Min J, Huang K, Shi C, Li L, Li F, Zhu T, and Deng H, et al. 2020. Pulmonary Cryptococcosis: comparison of Cryptococcal antigen detection and radiography in Immunocompetent and Immunocompromised patients. BMC Infectious Diseases, 20(1): 91.
Muller HE, and Sethi KK. 1972. Proteolytic activity of Cryptococcus neoformans against human plasma proteins. Medical Microbiology and Immunology, 158: 129–134.
Muselius B, Durand S, and Geddes-McAlister J. 2021. Proteomics of Cryptococcus neoformans : From the Lab to the Clinic. International Journal of Molecular Sciences, 22(22): 12390.
O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, and Alspaugh JA, et al. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase a regulates capsule. PLoS Pathogens.
Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, and Huffnagle GB, et al. 2004. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. American Journal of Pathology.
Ost KS, O’Meara TR, Huda N, Shannon KE, and Alspaugh JA. 2015. The Cryptococcus neoformans Alkaline response pathway: identification of a Novel Rim pathway activator. PLOS Genetics, 11(4): e1005159.
Paharik AE, Kotasinska M, Both A, Hoang T-MN, Büttner H, Roy P, Fey PD, Horswill AR, and Rohde H, et al. 2017. The metalloprotease SepA governs processing of accumulation- associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Molecular Microbiology, 103: 860–874.
Pearson WR. 2013. An introduction to sequence similarity (“Homology”) searching. Current Protocols in Bioinformatics, 0(3).
Perfect JR. 2017. The antifungal pipeline: a reality check. Nature Reviews Drug Discovery, 16(9): 603–616.
Poirier S, Mayer G, Benjannet S, Bergeron E, Marcinkiewicz J, Nassoury N, et al. 2008. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. The Journal of Biological Chemistry, 283(4): 2363–2372.
Pombejra SN, Salemi M, Phinney BS, and Gelli A. 2017. The metalloprotease, Mpr1, engages AnnexinA2 to promote the transcytosis of fungal cells across the Blood-Brain barrier. Frontiers in Cellular and Infection Microbiology, 7(June): 1–16.
Qu J, Zhang X, Lu Y, Liu X, and Lv X, et al. 2020. Clinical analysis in immunocompetent and immunocompromised patients with pulmonary cryptococcosis in western China. Scientific Reports, 10(1): 9387.
Rajasingham R, Smith RM, Benjamin JP, Jarvis JN, Govender NP, Chiller TM, et al. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. The Lancet Infectious Diseases, 17: 873–881.
Ratnikov BI,. Rozanov DV, Tanya PI, Baciu PG, Zhang H, DiScipio RG, Chestukhina GG, et al. 2002. An alternative processing of integrin αv subunit in tumor cells by membrane type-1 matrix metalloproteinase. Journal of Biological Chemistry, 277(9): 7377–7385.
Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, and Finn RD. 2018. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Research, 46(D1): D624–D632.
Reinhard FBM, Eberhard D, Werner T, Franken H, Childs D, Doce C, Savitski MF, et al. 2015. Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nature Methods, 12(12): 1129–1131.
Retanal C, Ball B, and Geddes-McAlister J. 2021. Post-translational modifications drive success and failure of fungal–host interactions. Journal of Fungi, 7(2): 124.
Rodrigues ML, dos Reis FCG, Puccia R, Travassos LR, and Alviano CS, et al. 2003. Cleavage of human fibronectin and other basement membrane-associated proteins by a Cryptococcus neoformans serine proteinase. Microbial Pathogenesis, 34: 65–71.
Santangelo R, Zoellner H, Sorrell T, Wilson C, Donald C, Djordjevic J, Shounan Y, and Wright L, et al. 2004. Role of extracellular phospholipases and mononuclear phagocytes in dissemination of cryptococcosis in a murine model. Infection and Immunity, 72(4): 2229–2239.
Santi L, Beys-da-Silva WO, Berger M, Calzolari D, Guimarães JA, Moresco JJ, and Yates JR, et al. 2014. Proteomic profile of Cryptococcus neoformans biofilm reveals changes in metabolic processes. Journal of Proteome Research, 13(3): 1545–1559.
Santiago-Tirado FH, Onken MD, Cooper JA, Klein RS, and Doering TL, et al. 2017. Trojan horse transit contributes to blood-brain barrier crossing of a eukaryotic pathogen. mBio, 8(1).
Sethi G, Shanmugam MK, Warrier S, Merarchi M, Arfuso F, Prem Kumar A, and Bishayee A, et al. 2018. Pro-apoptotic and anti-cancer properties of diosgenin: a comprehensive and critical review. Nutrients, 10(5): 645.
Shi M, Colarusso P, and Mody C. 2012. Real-time in vivo imaging of fungal migration to the central nervous system. Cell Microbiology, 14: 1819–1827.
Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, and Mody CH, et al.2010. Real-time imaging of trapping and urease- dependent transmigration of Cryptococcus neoformans in mouse brain. The Journal of Clinical Investigation, 120(5): 1683–1693.
Sidrim JJC, Perdigão-Neto LV, Cordeiro RA, Brilhante RSN, Leite JJG, Teixeira CEC, et al. 2012. Viral protease inhibitors affect the production of virulence factors in Cryptococcus neoformans. Canadian Journal of Microbiology, 58(7): 932–936.
Siscar-Lewin S, Hube B, and Brunke S. 2019. Antivirulence and avirulence genes in human pathogenic fungi. Virulence.
Sloan DJ, and Parris V. 13 May 2014. Cryptococcal meningitis: Epidemiology and therapeutic options. Clinical Epidemiology. 6: 169–182.
Smith KD, Achan B, Hullsiek KH, McDonald TR, Okagaki LH, Alhadab AA, et al. 2015. Increased antifungal drug resistance in clinical isolates of Cryptococcus neoformans in Uganda. Antimicrobial Agents and Chemotherapy, 59: 7197–7204.
Sorrell TC, Juillard P-G, Djordjevic JT, Kaufman-Francis K, Dietmann A, Milonig A, Combes V, and Grau GER, et al. 2016. Cryptococcal transmigration across a model brain blood-barrier: evidence of the Trojan horse mechanism and differences between Cryptococcus neoformans var. grubii strain H99 and Cryptococcus gattii strain R265. Microbes and Infection, 18(1): 57–67.
Steen BR, Lian T, Zuyderduyn S, MacDonald WK, Marra M, Jones SJM, and Kronstad JW, et al. 2002. Temperature-Regulated Transcription in the Pathogenic Fungus Cryptococcus neoformans. Genome Research, 12: 1386–1400.
Thangadurai D, Viswanathan MB, and Ramesh N. 2002. Indigoferabietone, a novel abietane diterpenoid from Indigofera longeracemosa with potential antituberculous and antibacterial activity. Die Pharmazie, 57(10): 714–715.
van Duin D, Casadevall A, and Nosanchuk J. 2020. Melanization of Cryptococcus neoformans and Histoplasma capsulatum reduces their susceptibilities to amphotericin B and caspofungin. Antimicrobial Agents and Chemotherapy, 46: 3394–3400.
Vu K, Eigenheer RA, and Phinney BS. 2013. Cryptococcus neoformans promotes its transmigration into the central nervous system by inducing molecular and cellular changes in Brain Endothelial cells. Infection and Immunity, 81(9): 3139–3147.
Vu K, Garcia JA, and Gelli A. 2019. Cryptococcal meningitis and antivirulence therapeutic strategies. Frontiers in Microbiology, 10(353).
Vu K, Tham R, Uhrig JP, Thompson GR, Pombejra SN, Jamklang M, Bautos JM, Gelli A, et al. 2014. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. mBio, 5(3).
Wang Y, and Casadevall A. 1994. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infection and Immunity, 62: 3004–3007.
Winter MB, Salcedo EC, Lohse MB, Hartooni N, Gulati M, Sanchez H, et al. 2016. Global identification of biofilm-specific proteolysis in Candida albicans. MBio, 7(5): e01514–16.
Xu C, Zhu H-M, Wu J-H, Wen H, and Liu C-J, et al. 2014. Increased permeability of blood – brain barrier is mediated by serine protease during Cryptococcus meningitis. Journal of International Medical Research, 42(1): 85–92.
Yoneda A, and Doering TL. 2006. A Eukaryotic Capsular Polysaccharide Is Synthesized Intracellularly and Secreted via Exocytosis. Molecular Biology of the Cell.
Zaragoza O. 2019. Basic principles of the virulence of Cryptococcus. Virulence, 10(1): 490–501.
Zhu, Y., Aebersold R, Mann M, and Guo T, et al. 2021. SnapShot: clinical proteomics. Cell, 184(18): 4840–4840.e1.
Information & Authors
Information
Published In

FACETS
Volume 7 • Number 1 • January 2022
Pages: 319 - 342
Editor: Emile Gluck-Thaler
History
Received: 1 October 2021
Accepted: 9 December 2021
Version of record online: 3 March 2022
Notes
This paper is part of a collection titled “The Canadian Fungal Network 2021 Annual Meeting – Connecting fungal researchers across Canada”.
Copyright
© 2022 Gutierrez-Gongora and Geddes-McAlister. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
All conceptualized the topic.
DG-G prepared the first manuscript draft.
DG-G provided figures.
All edited and prepared the final manuscript version.
All have read and agreed to the published version of the manuscript.
Competing Interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of this manuscript, or in the decision to publish the results.
Funding Information
This work was supported, in part, by the University of Guelph, Canadian Foundation of Innovation (JELF 38798), New Frontiers Research Fund: Exploration, Banting Research Foundation—Jarislowsky Discovery Award, and Canadian Institutes of Health Research—Project Grant for JG-M
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Davier Gutierrez-Gongora and Jennifer Geddes-McAlister. 2022. Peptidases: promising antifungal targets of the human fungal pathogen, Cryptococcus neoformans. FACETS.
7(): 319-342. https://doi.org/10.1139/facets-2021-0157
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Antifungal Potential of Synthetic Peptides against Cryptococcus neoformans: Mechanism of Action Studies Reveal Synthetic Peptides Induce Membrane–Pore Formation, DNA Degradation, and Apoptosis
