Introduction
Pacific salmon (
Oncorhynchus spp.) are a group of anadromous species that experience extreme spatial variations in ecosystem attributes during their early marine life, during which they typically migrate from estuaries to the coastal ocean and then to the open ocean. Pacific salmon can experience high mortality during their first months at sea, with mortalities of up to 80-90% estimated for tagged salmon (
Welch et al. 2011). It has been hypothesized that growth and mortality rates during this phase may contribute to interannual variability and long-term population trends of salmon stocks (
Friedland et al. 2000;
Mueter et al. 2005). Although there are likely numerous factors that influence juvenile salmon survival during their early marine phase, correlative evidence suggests that feeding conditions play an important role (
Beamish et al. 2012;
Duffy and Beauchamp 2011). Through impacts on growth, feeding conditions have the potential to affect vulnerability to predation (
Gliwicz 2002), ability to survive stressful ocean conditions (
Lasker 1978), and the body condition required to survive their first ocean winter (
Beamish and Mahnken 2001).
Recent research from North America’s west coast has drawn attention to the potential influence of small-scale coastal marine conditions on juvenile salmon survival (
Ferriss et al. 2014;
Hunt et al. 2018;
Journey et al. 2018;
McKinnell et al. 2014). Out-migrating juvenile Fraser River sockeye (
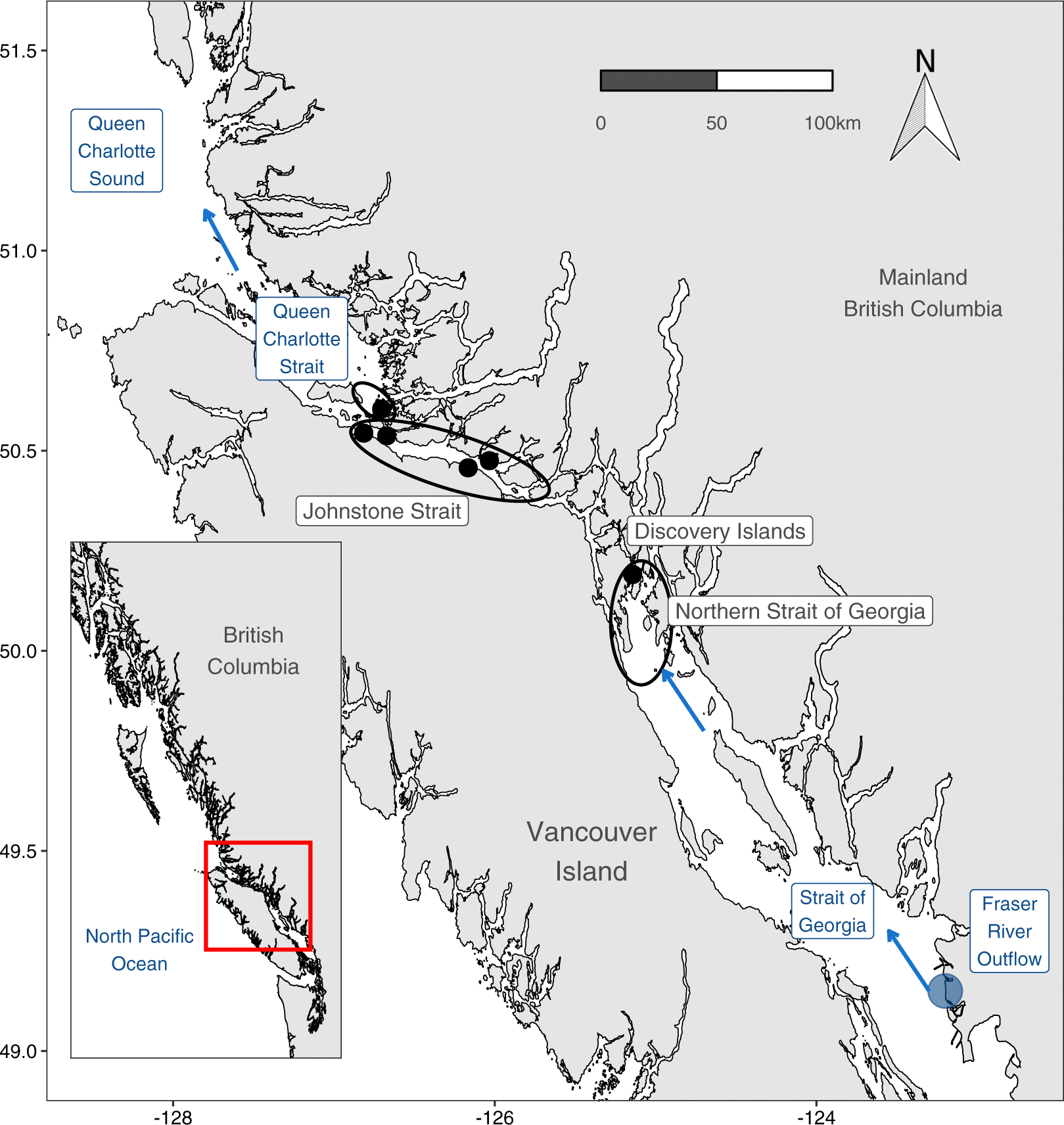
Oncorhynchus nerka) first enter the ocean into the stratified, comparatively warm, and highly productive Strait of Georgia (SoG) basin of the Salish Sea (
Fig. 1). Most of these juveniles migrate northwards through the SoG into the tidally mixed Discovery Islands (DI) and Johnstone Strait (JS) before reaching the productive Queen Charlotte Strait (QCS) (
McKinnell et al. 2014).
Thomson et al. (2012) hypothesized that poor foraging conditions for juvenile salmon in the SoG and QCS before entering the Gulf of Alaska were responsible for high mortality and low adult returns for that year class. In the Trophic Gauntlet Hypothesis (TGH),
McKinnell et al. (2014) went further to propose that these anomalous conditions in QCS and SoG were amplified by consistently poor foraging conditions in JS, where strong tidal mixing causes light limited primary production and low zooplankton biomass. Subsequent research has demonstrated that tidally mixed waters in JS are indeed characterized by poor foraging conditions, with a prevalence of small zooplankton (< 2 mm;
Mahara et al. (2021)), corresponding with low foraging success for migrating juvenile sockeye salmon (
James et al. 2020). Low insulin-like growth factor 1 (IGF-1) levels in JS compared to higher levels in the northern Strait of Georgia (NSoG), QCS and Queen Charlotte Sound supports an impact of these foraging conditions on juvenile salmon growth (
Ferriss et al. 2014;
Journey et al. 2018).
Relating growth proxies to environmental conditions needs to consider the integration time of the proxy used. IGF-1 is a biochemical proxy of growth that has been shown to integrate feeding conditions experienced over 4–14 days (
Beckman et al. 2004), relatively long when assessing the response of fast migrating organisms over short distances (
Duguid et al. 2018;
Gabillard et al. 2006;
Pierce et al. 2005). The need for precise handling when sampling in the field, and the difficulty in sampling from individuals smaller than 120 mm also make the IGF-1 method difficult to routinely apply in juvenile salmon research (
Duguid et al. 2018;
Ferriss et al. 2014). Another growth proxy commonly used in fish ecology and fishery research is RNA/DNA ratio (
Clemmesen and Doan 1996). The underlying principle of using the ratio of RNA to DNA is that the concentration of DNA per cell is constant, whereas the RNA concentration per cell varies with its anabolic activity. The RNA/DNA ratio integrates feeding history over a period of 1–5 days (
Buckley et al. 1999;
Clemmesen and Doan 1996;
Wright and Martin 1985), which makes it useful for assessing fish response to rapid spatial changes in coastal migration habitat. RNA/DNA can be effectively sampled irrespective of individual size and can be sampled from individuals frozen in the field without any degradation.
Another variable that needs to be considered when assessing fish growth response is foraging conditions. Prey quantity is often considered one of the main drivers of growth, survival, and abundance of marine organisms (
Bilton and Robins 2011), however, prey quality also plays a critical role (
Litz et al. 2017). Fatty acids (FA), and essential fatty acids (EFA) in particular, are useful indicators of both food quality and the nutritional condition of a fish (
Tocher 2003). EFAs are not synthesised
de novo by vertebrates in sufficient quantities to meet their physiological demands (
Xu et al. 2018). Rather, EFAs are primarily synthesised by phytoplankton and subsequently reach consumers through trophic transfer (
Dalsgaard et al. 2003). Marine carnivorous fish, such as sockeye salmon, have only a limited ability to convert EFAs into important long-chained PUFAs such as docosahexaenoic acid (DHA; 22:6n-3), eicosapentaenoic acid (EPA; 20:5n-3), and therefore must be obtained from their diets (
NRC 2011). Studies have shown that DHA and EPA concentrations measured in fish muscle tissue reflect the FA composition of their diets (
Jin et al. 2017;
Xu et al. 2020). Total amounts of DHA and EPA are known to affect the development, growth, and survival of juvenile marine fish (
Mourente et al. 1991) and are of major importance during periods of rapid growth (
Jin et al. 2017) (
Table 1). Low levels of DHA and EPA can lead to reduced growth, maldevelopment, reduced reproduction, and ultimately decreased survival (
Copeman and Parrish 2002) (
Table 1). Stored FAs, especially DHA and EPA, are also important when fish experience times of food limitation and starvation, for energy mobilization, hyperphagia, and for compensatory growth.
Experimental studies have shown that food quality changes, especially micronutrient limitation, and associated fatty acid changes lead to reduced RNA/DNA as lipid and protein metabolism is compromised when supply is limited (
Boersma et al. 2008). The fatty acid composition integrates the feeding history over a period of approximately one week in juvenile Chinook salmon (
Garzke et al. 2022), and using RNA/DNA and fatty acids in combination therefore provides a useful approach to identify changes in nutritional and physiological condition in organisms (
Clemmesen 1994;
John et al. 2001;
Malzahn et al. 2007;
Paulsen et al. 2014;
Xu et al. 2009)
In this study, we examined the response of juvenile sockeye salmon RNA/DNA, fatty acids and body mass index as they migrated through complex oceanographic habitats in coastal British Columbia, Canada. Specifically, our study area spanned the trophic gauntlet region (
McKinnell et al. 2014) allowing assessment of the fish nutritional condition when migrating through JS, a region previously demonstrated to provide food poor conditions.
Discussion
Juvenile salmon encounter diverse ocean conditions during their early marine life. In this study, we compared the body condition of juvenile salmon (RNA/DNA) to their nutritional condition (fatty acid profiles) and prey availability (zooplankton biomass) across an ∼ 120 km section of their migration route in coastal British Columbia, in 2015 and 2016. Both body and nutritional condition of juvenile sockeye salmon showed short term changes that differed between years but were not related to zooplankton biomass, abundance, average size, or species composition. Our RNA/DNA data showed that the recovery of juvenile salmon in QCS after a period of starvation in JS might depend on their condition prior to entering JS. Furthermore, our study provides evidence that the Trophic Gauntlet Hypothesis should be extended to include food quality (e.g., fatty acid content), not just food quantity. Below we discuss the role of fish condition in areas with coastal oceanographic variation, and the cumulative effect of fish encountering a mosaic of habitats during migration.
As outlined in the introduction, the majority of out-migrating juvenile Fraser River sockeye salmon travel northwards through the SoG before entering a section of tidally mixed channels in the DI and JS (
Furey et al. 2015). In the SoG, they typically encounter surface waters that are relatively warm, fresh, and stratified, correlated with high phytoplankton biomass, high zooplankton biomass, and high mean zooplankton size (
James et al. 2020;
Mahara et al. 2021). Ocean conditions become cooler and saltier as the fish move north into the DI, JS, and QCS. This change in ocean conditions is driven by the deep tidal mixing that brings cold and salty deep water to the surface (
McKinnell et al. 2014). Phytoplankton biomass is low through the tidally mixed northern DI and JS, leading to both low zooplankton biomass and smaller zooplankton size. This pattern is, however, reversed in QCS.
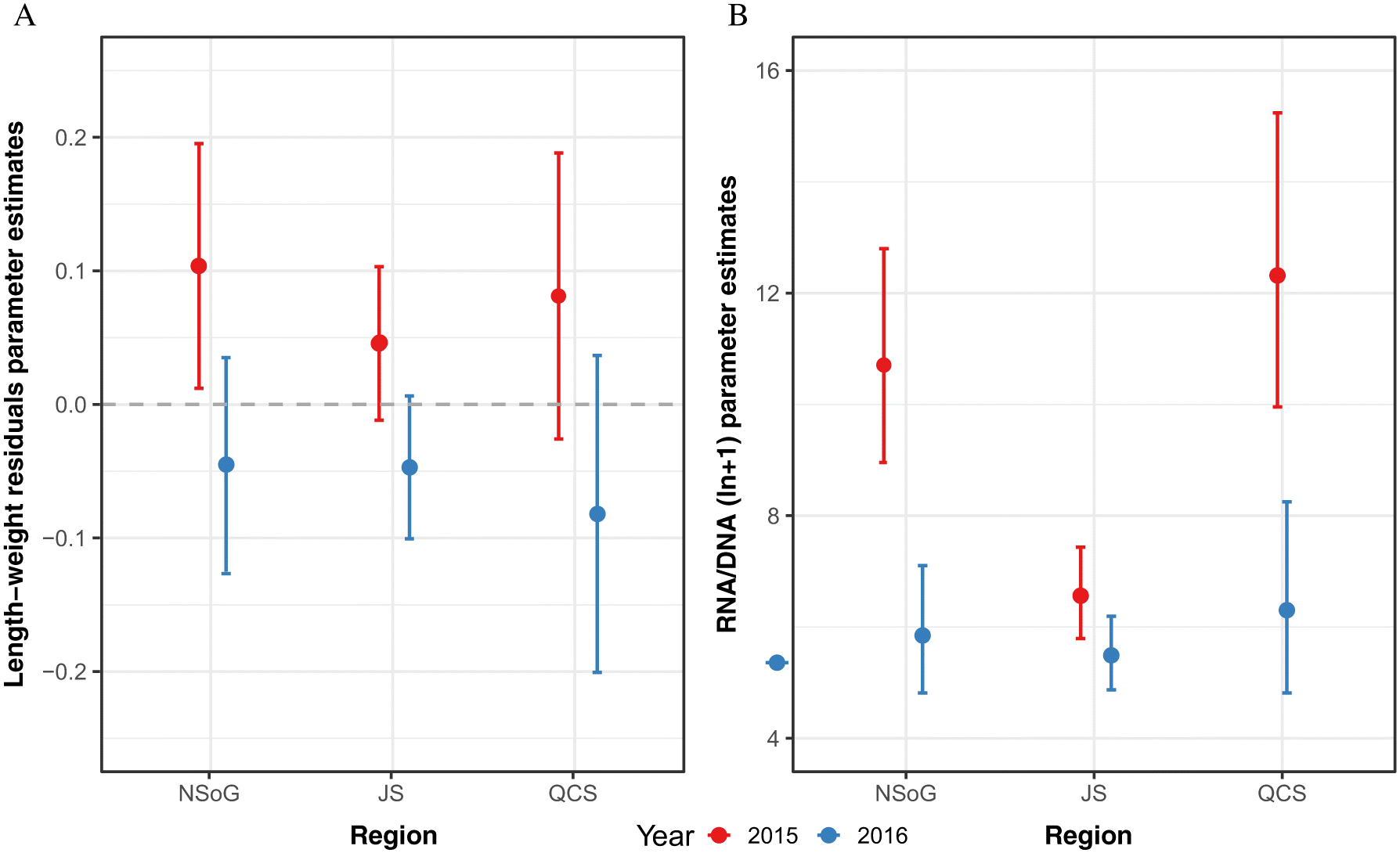
In the present study, juvenile salmon sampled in 2015 had RNA/DNA ratios which supported the Trophic Gauntlet Hypothesis (
McKinnell et al. 2014), with high fish condition in the NSoG and QCS, and low condition in JS. The link between RNA/DNA and feeding success is supported by juvenile sockeye salmon stomach fullness analysis across the study area in 2015, which found high stomach fullness in the NSoG (Gut Fullness Index (GFI) = 1%–2%), low fullness in DI (GFI = 0.2 %), low fullness in JS (GFI = 0.48 %), and full stomachs in QCS (GFI = 3.5%) (
James et al. 2020). Poor foraging conditions for juvenile sockeye salmon, with GFI < 1, were also observed in 2016 (
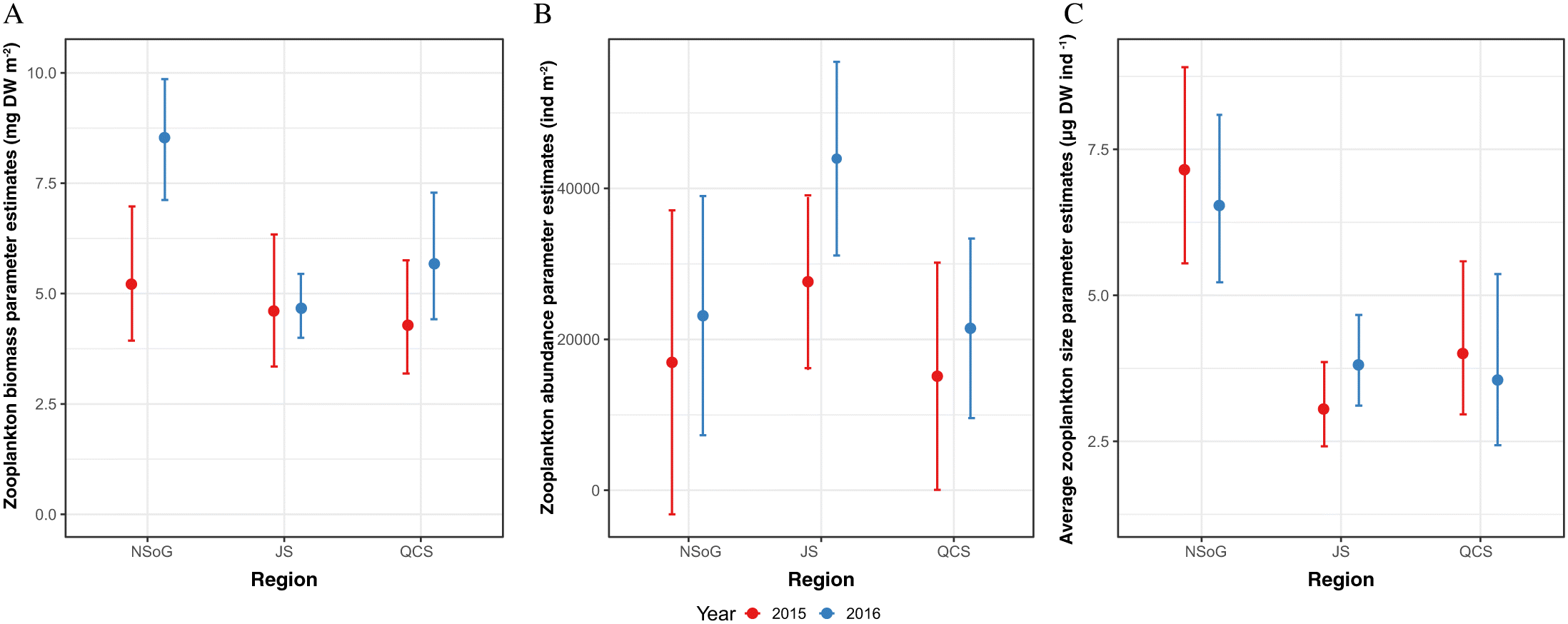
James 2019). Length-weight residuals showed a weaker response over the migration route, likely reflecting their longer integration times and highlighting the importance of more sensitive growth proxies, with faster turnover, to understand the response of fish to small scale regional changes in environmental conditions. Notably, RNA/DNA ratios and length-weight residuals showed a significantly different pattern in 2016. RNA/DNA ratios and length-weight residuals were significantly lower in NSoG juvenile salmon compared to 2015, indicating that feeding conditions in the NSoG were poorer in 2016 than 2015. In 2016, RNA/DNA ratios remained low during the migration through JS and increased only slightly in QCS. The lower condition in 2016 occurred despite similar zooplankton biomass, abundance, and average size to 2015, indicating that another factor was contributing to juvenile salmon nutritional and body condition.
There was no correlation between RNA/DNA and SST, indicating that temperature did not lead to the interannual difference in body condition. However, there were significant differences in juvenile sockeye salmon fatty acid profiles both between regions and years. Stored lipids and fatty acids in fish tissues are important to mobilize energy from muscle, liver, and viscera by FA β-oxidation processes during periods of starvation (
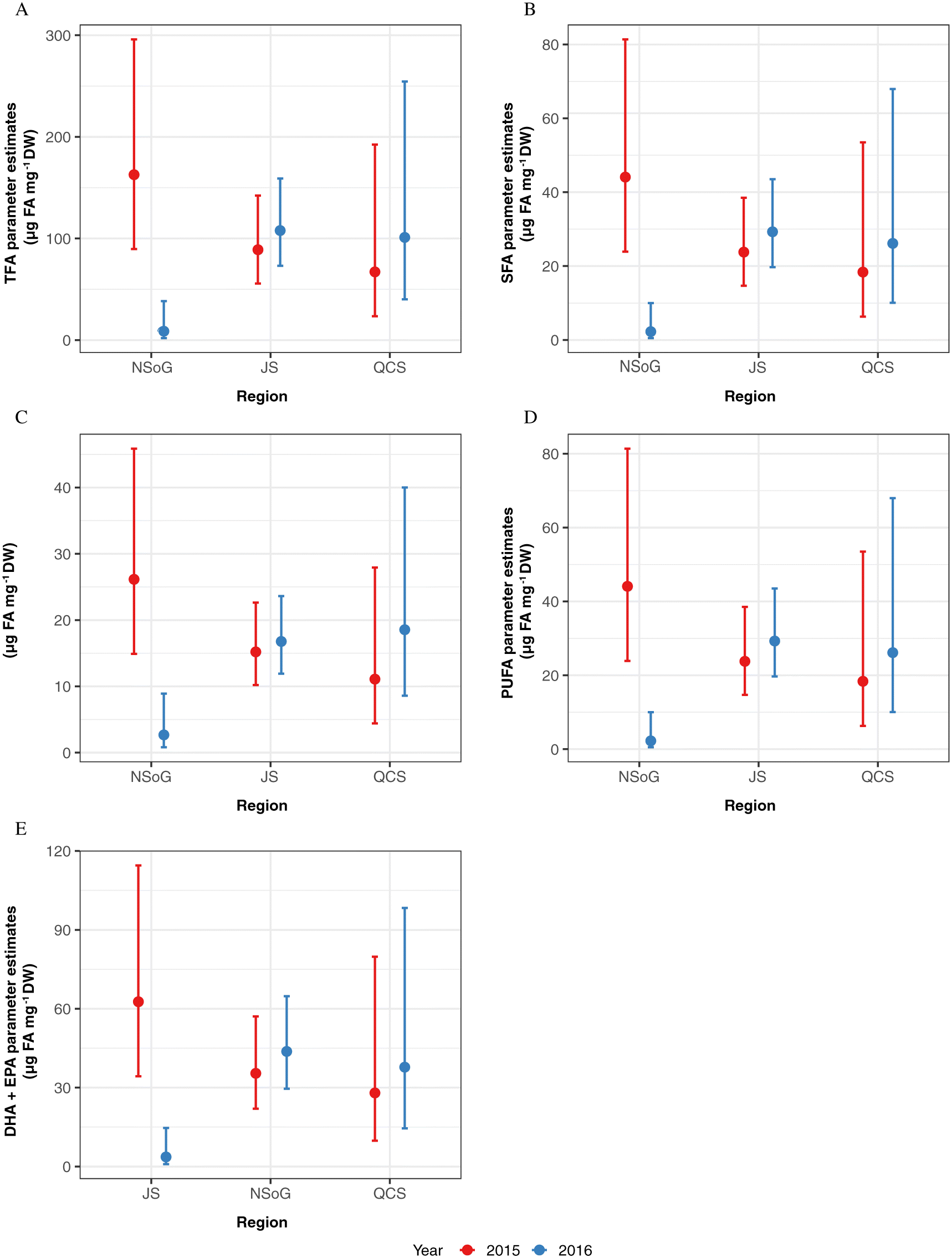
Halver and Hardy 1989). During starvation, saturated FA (SFA) are the first FAs to be mobilized for ATP production, followed by monounsaturated FA (MUFA) and then polyunsaturated FA (PUFA) (
Wen et al. 2006). In 2015, we detected a decrease of TFA, SFA, MUFA, and PUFA along the migration corridor, with lowest concentrations in JS, indicating that fish had oxidized FAs stored in white muscle tissue for ATP production. In 2016, juvenile salmon sampled in the NSoG had significantly lower concentrations of TFA, SFA, MUFA, and PUFA than 2015, indicating that the nutritional condition of salmon was already very low when they reached the NSoG, with less FAs stored in white muscle tissue as an energy reserve for starvation periods.
As FAs have a response time of a minimum of one week, the condition of juvenile salmon sampled in the NSoG in this study must have been in response to the preceding conditions in the SOG. Differences between years in body and nutritional condition may have resulted because either food quantity or food quality were low in the SoG in 2016, leading to depletion of FA reserves needed to meet the energy demands of migration. Zooplankton biomass in the study area was not significantly different between 2015 and 2016 (this study;
Mahara et al. (2019)) and was significantly higher in the central SoG in 2016 (
Perry et al. 2021). This suggests that lower food quality (e.g., DHA + EPA) in 2016 was the most likely driver of lower fish FA content and RNA/DNA ratios in that year. As outlined in the introduction, low DHA and EPA availability in prey have important implications for fish condition, including reduced growth rates, visual maldevelopment with negative effects on hunting efficiency, an increase in FAs that are considered as pro-inflammatory (
Table 1), and reduced capacity for compensatory growth, i.e., accelerated growth that follows a period of limited growth once non-limiting conditions are encountered (
Ballantyne et al. 2003;
Bou et al. 2017).
Fish obtain DHA and EPA through their diet. In the case of zooplankton prey, variations in DHA and EPA can occur because of shifts in composition, with different species having different nutritional quality and fatty acid content (
Hiltunen et al. 2021), or shifts in the nutrition of phytoplankton that the zooplankton feed on (
Costalago et al. 2020;
El-Sabaawi et al. 2009). There is substantial variability in FA content among zooplankton species in the Salish Sea (
El-Sabaawi et al. 2009;
Hiltunen et al. 2021). However, analysis of zooplankton communities has shown that species composition in the NSoG did not differ between 2015 and 2016 (
Mahara et al. 2019), indicating that this was not an important factor in zooplankton prey quality. It should be noted that similarly, there was no interannual difference in zooplankton community composition in the DI, JS, or QCS either (
Mahara et al. 2021). With regard to zooplankton’s phytoplankton prey, no major differences were observed in phytoplankton composition in the NSoG between 2015 and 2016 (
Belluz et al. 2021). However, spring bloom timing differed substantially between years: in 2015 an unusually early spring bloom (February 24th) occurred, while in 2016 the bloom timing was typical (April 1st) with maximum biomass of 12.40 mg Chl
a m
−3 and 11.53 mg Chl
a m
−3, respectively (
Belluz et al. 2021;
Mahara et al. 2019). It is possible that the later phytoplankton bloom in 2016 delayed the development of the zooplankton community and onset of lipid accumulation in the later life stages (e.g.,
Mayzaud et al. (1999))
The better body and nutritional condition of juvenile salmon leaving the NSoG in 2015 appeared to effect recovery after leaving JS. In 2015, juvenile salmon RNA/DNA ratios in QCS showed compensatory growth, almost doubling after passing JS where RNA/DNA ratios were low and FA concentrations declined due to necessary energy mobilization. In contrast, in 2016 when juvenile salmon already had low condition and depleted energy reserves in the NSoG, no compensatory growth was observed in QCS. Studies with Atlantic salmon (
Salmo salar) have shown that compensatory growth success depends on the quality of food available, particularly requiring high lipid content (
Johansen et al. 2001).
Turchini et al. (2007) proposed the term ‘lipo-compensatory growth’ after showing that Murray cod were able to compensate for starvation and refuel fatty acid storage when fed with high fatty acid diets. This supports the importance of high-quality food for enduring periods of poor foraging conditions.
Protracted periods of poor foraging may not only slow down recovery but may also have long-term impacts on the physiology of the fish (
Nikki et al. 2004). Periods with very low food biomass longer than 3–4 weeks can decrease digestive enzyme activity, inhibiting the ability of fish to sufficiently digest food when available again, resulting in overall lower body size and higher mortality (
Abolfathi et al. 2012). This is highly relevant to juvenile salmon migrating through the BC coastal ocean. The migration of juvenile sockeye salmon through the DI and JS takes approximately 14 days, while time spent in the SoG is estimated to be between 30 and 50 days (
Preikshot et al. 2012). Thus, as suggested by
McKinnell et al (2014), anomalously poor foraging conditions in the SoG and (or) QCS, coupled with typically poor foraging conditions in the DI and JS, may have an additive effect that impacts early marine survival of juvenile salmon. Our study goes further, providing evidence that the Trophic Gauntlet Hypothesis should be extended to food quality as a critical factor in juvenile salmon growth and survival.