Amphibian ranaviruses in Europe: important directions for future research
Abstract
Ranaviruses are an emerging group of pathogens capable of infecting all cold-blooded vertebrates. In Europe, ranaviruses pose a particularly potent threat to wild amphibian populations. Since the 1980s research on amphibian-infecting ranaviruses in Europe has been growing. The wide distribution of amphibian populations in Europe, the ease with which many are monitored, and the tractable nature of counterpart ex situ experimental systems have provided researchers with a unique opportunity to study many aspects of host–ranavirus interactions in the wild. These characteristics of European amphibian populations will also enable researchers to lead the way as the field of host–ranavirus interactions progresses. In this review, we provide a summary of the current key knowledge regarding amphibian infecting ranaviruses throughout Europe. We then outline important areas of further research and suggest practical ways each could be pursued. We address the study of potential interactions between the amphibian microbiome and ranaviruses, how pollution may exacerbate ranaviral disease either as direct stressors of amphibians or indirect modification of the amphibian microbiome. Finally, we discuss the need for continued surveillance of ranaviral emergence in the face of climate change.
Introduction
In 1979 a Scandinavian population of captive Atlantic cod (Gadhus morhua) began to exhibit elevated levels of morbidity and mortality. Affected fish exhibited a novel condition characterised by severe skin ulcerations, and investigation of the causes behind this disease outbreak implicated a virus belonging to the group Iridoviridae (Jensen et al. 1979). Since the advent of modern molecular techniques, researchers have revisited this case and genomic analysis of the isolated viruses has shown that this outbreak was, in fact, the first recorded incidence of ranavirosis (the often lethal disease caused by ranaviruses) in Europe (Ariel et al. 2010). Despite the importance of aquaculture and fisheries to many European economies (Crilly and Esteban 2013), the viral diversity and host range of ranaviruses infecting farmed and wild fishes in Europe has received only sporadic research effort over the last four decades (see recent reviews by Price et al. 2017b; Allain and Duffus 2019). However, fish are not the only group of vertebrates impacted by ranavirosis in Europe; ranaviruses are also known to establish infections in European herpetofauna (Cunningham et al. 1997; Balseiro et al. 2009). To date, reports of ranaviral infection and disease in reptiles are rare (Marschang 2011; Price et al. 2014, 2017b), but ranavirosis is considered to be an emerging infectious disease of European amphibians and is believed to pose a significant conservation threat (Daszak et al. 1999; Earl and Gray 2014; Campbell et al. 2018a).
The first incidences of ranavirosis in European amphibians were reported by members of the public in the United Kingdom during the late 1980s (Cunningham et al. 1997). Since that time, research effort devoted to these deadly pathogens has slowly grown both globally and in Europe (Fig. 1). It is currently accepted that there are two primary lineages of ranavirus circulating within European amphibians, the Frog Virus 3 (FV3)-like and the common midwife toad virus (CMTV)-like viruses (reviewed in Price et al. 2017a). Although there is overlap in the geographical range of these viruses, each is apparently impactful in a particular continental region, separating the study of amphibian ranaviruses in Europe into two distinct theatres: continental Europe and the islands that comprise the United Kingdom. The distribution and severity of ranavirus outbreaks in Europe have been given in-depth treatment in two recent reviews (Price et al. 2017a; Allain and Duffus 2019). Our goal in this article is not to supplant those works. Instead, here we highlight gaps in our knowledge concerning the forces shaping interactions between amphibian hosts and ranaviruses, and predictors of the severity of ranavirosis outbreaks, that should be prioritised for research. First, we present a brief summary of the key research findings from each of the main European theatres, before highlighting important questions that remain to be answered and potential avenues of future investigation.
Fig. 1.
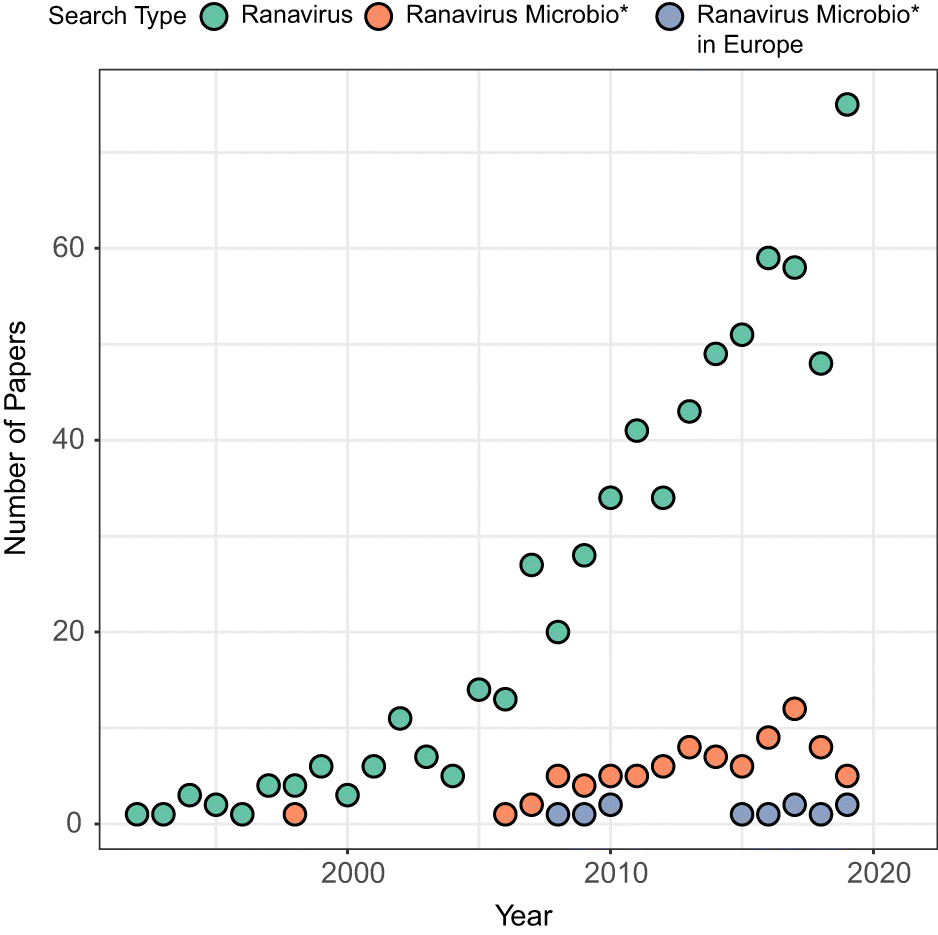
Ranavirosis in the United Kingdom
The United Kingdom is densely populated, resulting in significant overlap between the human population and wildlife (Fuller and Gaston 2009). As a result, many United Kingdom amphibian populations reside in small water bodies, such as urban garden ponds, where they are easily observed by humans. In the late 1980s members of the British public began to report unusual mass mortality in such populations of the European common frog (Rana temporaria; Cunningham et al. 1997). Deceased adult R. temporaria were found exhibiting severe skin ulceration and hemorrhaging of the internal organs (Cunningham et al. 1997). The Frog Mortality Project (FMP) was established in 1992 by scientists at the Zoological Society of London to collate and investigate these reports of disease incidence (Cunningham et al. 1997). The FMP solicited reports of frog mortality from the general public using an outreach campaign and facilitated the collection and post-mortem examination of carcasses collected at the site of reported outbreaks. Pathological and histological examination revealed that a ranavirus closely resembling FV3 was the likely cause (Cunningham et al. 1997). Comparative molecular analysis showed that this virus was highly similar to North American isolates of FV3 and was likely introduced into the United Kingdom via the international trade in amphibians and fish (Hyatt et al. 2000).
The FMP has generated a large archive of both amphibian tissue samples and associated viral isolates. Genomic analyses of ranavirus isolates collected as part of the FMP have shown that not only were there at least two distinct introduction events of FV3 into the United Kingdom (Price et al. 2016, 2017a; Duffus et al. 2017) but also that an additional ranaviral species, CMTV, has been present in the United Kingdom since as early as 1995 (the earliest known detection of CMTV; Price et al. 2017a). Despite the presence of CMTV, it is believed that all records of ranavirosis in the United Kingdom are associated with FV3 (Price et al. 2016, 2017a). In stark contrast to ranavirosis outbreaks caused by FV3 elsewhere in the world, which often exhibit an extensive host range in terms of species and life stages impacted, mortality due to ranavirosis in the United Kingdom is almost entirely limited to R. temporia (Duffus et al. 2013; Allain and Duffus 2019) and is exclusively observed in post-metamorphic animals (Duffus et al. 2013).
Through the collection of disease incidence records the FMP has also spawned a database of R. temporaria populations that reside on private property and whose health is continually visually monitored by property owners (Teacher et al. 2010; Lawson et al. 2015). With the help of these “citizen scientists” researchers in the United Kingdom have been able to establish a network of R. temporaria populations of known disease history, allowing for several comparative and (or) longitudinal studies that have advanced our understanding of interactions between ranaviruses and their hosts at the individual and the population level (Lawson et al. 2015). Research within this network of comparative populations has shown that the emergence of ranavirosis has resulted in a reduction of R. temporaria numbers by an average of over 80% in infected populations, though local extinction and recovery have also been documented (Teacher et al. 2010). Additionally, ranavirosis appears to have (i) altered the population genetics (Teacher et al. 2009a), (ii) reversed the mating system (Teacher et al. 2009b); and (iii) truncated the age structure of impacted R. temporaria populations (Campbell et al. 2018a). Applying predictive models to disease incidence reports and their associated environmental metadata has also revealed that the spread of FV3 within the United Kingdom is very likely facilitated by human translocation and pathogen pollution (Price et al. 2016) and that frequency and severity of ranavirosis outbreaks is tightly linked to the climactic conditions (Price et al. 2019). Data drawn from these incidence reports have also revealed that the occurrence and severity of ranavirosis outbreaks are correlated with host population density, the presence of secondary host species (hosts which can become infected with and shed virus but are themselves asymptomatic, including the common toad (Bufo bufo) and various species of fish), and the use of garden chemicals (North et al. 2015). Epidemiological models have demonstrated that the persistence of ranaviruses within R. temporaria populations can be maintained via exclusively adult to adult contact transmission (Duffus et al. 2019); however, the drivers of the distribution and intensity of ranavirus infection in the United Kingdom remain poorly understood. Though many monitored common frog populations in the United Kingdom appear to have never suffered outbreaks of ranavirosis, recent research detected the presence of FV3-like ranavirus in frogs originating from all populations studied, including those with no history of observable ranavirosis (Campbell et al. 2018b). Critically, this suggests that ranaviruses may be ubiquitously distributed within R. temporaria populations in the United Kingdom, but that whether or not a population develops ranavirosis depends on some additional cryptic factors that promote infection of hosts and cause those infections to reach clinical or lethal thresholds.
For example, there is growing evidence suggesting a potential link between the occurrence of ranavirosis and composition of bacterial communities residing on the amphibian skin. Differing bacterial communities have been detected on the skin of frogs originating from populations with endemic ranavirosis versus frogs from populations with no history of disease, based on both transcriptomic (Campbell et al. 2018b) and genomic DNA (Campbell et al. 2019) sequences. Beyond these field-based studies, the outcome of acute ranaviral infection has been shown to be impacted by the diversity of the amphibian skin microbiome during an experimental study that showed that metamorphic R. temporaria with a depauperate bacterial microbiome were more likely to succumb to infection with FV3 than were metamorphic R. temporaria possessing a skin microbiome with higher species richness (Harrison et al. 2019).
Ranavirosis on continental Europe
Despite a long history of ranaviruses impacting fish on continental Europe (Jensen et al. 1979; Ariel et al. 2010), since mid-2000 ranavirosis has also been emerging in amphibians. The primary causes of ranavirus-induced mortality on continental Europe appear to be several strains of CMTV (reviewed by Price et al. 2017b). CMTV are considered a distinct species complex within the genus Ranavirus (Chinchar et al. 2017) and were first discovered during investigations of a large mortality event that occurred at the Picos de Europa National Park, Spain, in 2007 (Balseiro et al. 2009). This outbreak impacted the larval life-stage of the common midwife toad (Alytes obstetricans), lending the virus its name. Since 2007 there have been repeated outbreaks of ranavirosis attributed to CMTV-like ranaviruses throughout continental Europe, affecting a wide range of species and life stages. Mass mortality events have been observed in Danish edible frogs (Pelophylax kl. Esculentus; Ariel et al. 2009) and a French population of R. temporaria (Miaud et al. 2016). Worryingly, CMTV has also been documented to cause local outbreaks of ranavirosis that can span entire amphibian community assemblages and all life stages, causing pronounced and persistent population declines (Price et al. 2014; Rijks et al. 2016; Rosa et al. 2017).
Several strains of CMTV are known to be circulating in Europe, and experimental work has revealed these strains to vary markedly in pathogenicity (Saucedo et al. 2018, 2019). A lack of genetic diversity in CMTV isolates collected from various outbreaks and a high number of genetic loci that appear to be under strong selection has led to the hypothesis that CMTV is an invasive pathogen on continental Europe (Price 2015; Price et al. 2017b), though the route of invasion remains unknown. Although FV3-like ranaviruses are known to occur in Europe in both amphibians and reptiles (Price et al. 2014; Stöhr et al. 2015) and are known to occur in sympatry with CMTV-like viruses, such incidences are rare (Price et al. 2017b). Recent studies have shown that in regions where both viral species groups co-occur there is potential for high levels of recombination resulting in hybrid viruses, which could potentially possess elevated virulence compared to either CMTV- or FV3-like viruses (Price et al. 2014; Price 2015; Claytor et al. 2017; Rosa et al. 2017). Overlap also exists between CMTV and the amphibian chytrid fungi, and co-infections are known to establish (Rosa et al. 2017); however, little evidence currently exists that either pathogen exacerbates the impact of the other (Rosa et al. 2017). The gross pathology of ranavirosis caused by CMTV is identical to that which is caused by FV3 (e.g., Cunningham et al. 1997; Balseiro et al. 2009); however, CMTV is known to replicate initially in the oral cavity of infected amphibians, before invading the connective tissues and subsequently the organs, including the skin (Saucedo et al. 2019). Research that focused on an alpine population of R. temporaria in France has also been key in proving the efficacy of environmental DNA or “e-DNA” at detecting the presence of ranaviruses in European amphibian populations (Miaud et al. 2019). Critically, the use of e-DNA not only allows for the detection of pathogens without the need for intensive or invasive sampling techniques of hosts, but also permits quantification of temporal change in pathogen loads in the environment (e.g., Hall et al. 2016; Miaud et al. 2019). Such data could prove invaluable for attempting to understand spatial variation in the severity of ranavirosis outbreaks.
Future directions
Although European research has been instrumental in advancing the study of amphibian infecting ranaviruses around the world, there remain significant gaps in our understanding that need to be addressed if effective management and mitigation strategies of these emerging pathogens are to be developed. Below, we outline these key directions of future research and how they may be pursued. Although we give special consideration to how these knowledge gaps may be addressed in European amphibian systems, these directions of research should be considered a priority of researchers globally.
Further investigation of the interaction between the amphibian microbiome and ranaviruses
There is substantial and growing evidence that the amphibian skin microbiome is vital to the ability of amphibians to resist or tolerate infection by pathogens (Harris et al. 2006, 2009; Becker et al. 2015). Although this evidence is primarily drawn from studies concerning amphibians infected with the chytrid fungi, including Batrachochytrium dendrobatidis (Bd) (Kueneman et al. 2016; Harrison et al. 2020) and B. salamandrivorans (Bates et al. 2019), recent research suggests that host-associated microbial communities in amphibians also represent a key component of their immune defense against ranaviruses. For example, skin microbiome structure has been found to correlate with population disease-status in the wild (Campbell et al. 2018b, 2019) and survival of acute ranaviral infection under laboratory conditions (Harrison et al. 2019). Furthermore, gut microbiomes of developing wood frogs (Rana sylvatica) have been shown to be critical modulators of later life resistance to ranavirus (Warne et al. 2019). These are potentially important findings as the ability of probiotic treatments to mitigate chytridiomycosis has been experimentally demonstrated (e.g., Kueneman et al. 2016) and similar treatments for ranavirosis would significantly advance amphibian conservation efforts. However, despite the clear importance of the host-associated microbial communities to emerging pathogens such as ranaviruses, several significant gaps in our knowledge remain that must be addressed before such probiotic treatments could be developed.
Generality of microbiome–pathogen interactions
To date, all evidence of a link between ranavirosis and the amphibian skin microbiome originates from within the United Kingdom R. temporaria field system outlined above, and studies on the interaction between gut microbiomes, host immunity, and ranavirus remain rare (e.g., Warne et al. 2019). Understanding the generality of these patterns in other host species and locations is a vital first step in quantifying the importance of amphibian microbial communities in shaping resistance or tolerance to ranaviruses. As such, further field and experimental studies of the amphibian microbiome in populations with varying ranaviral disease history, particularly in a diversity of vulnerable species outside of the United Kingdom, are a crucial first area of future research.
How should we measure host–microbe interactions?
Understanding the ecology of host-associated microbes, and how they influence host health is challenging in wild systems. Researchers face the choice of a variety of molecular methods for quantifying the composition of microbial communities, including those that simply yield information on the presence and relative abundance of microbial taxonomic groups (e.g., 16S rRNA amplicon sequencing), and those that also directly estimate functional properties of the microbiome (e.g., metagenomics, metatranscriptomics, and metabolomics (Harrison and Cameron 2020)). An outstanding issue is that the precision with which we can measure host–microbe interactions, and thus the inferences we make, depend on which of these methods we use. For example, Campbell et al. (2018b) profiled the R. temporaria microbiome using bacterial reads filtered from a transcriptomics study of the response of wild frog populations to endemic ranavirosis. They found that Bacillus subtilis, which is a bacterium commonly used as a probiotic in the aquaculture industry (e.g., Aly et al. 2008; Liu et al. 2012; Ran et al. 2012), was two orders of magnitude more abundant on frogs that were persisting at sites with endemic disease versus frogs from ranavirosis-free populations. However, the same pattern was not detected during a follow-up study that used 16S rRNA amplicon sequencing to categorise and compare the skin bacterial community structure of the same populations from samples collected the following year (Campbell et al. 2019). This discrepancy between results based on relative bacterial abundance versus bacterial transcription raises two interesting questions that warrant further study. Firstly, does transcriptional activity or relative biomass present the most informative indication of the importance of a microbial species in the amphibian skin microbiome? Understanding which potential measure is a truer reflection of genuine interaction between bacterial taxa and a pathogen will allow for more accurate detection of bacterial species which possess the potential to serve as probiotics in mitigation strategies. Targeted transcriptomic studies of the amphibian commensal skin bacteria (often termed the metatranscriptome) are rare (Rebollar et al. 2016). However, the incorporation of transcriptomic analysis in future research of the link between that amphibian skin microbiome and ranavirosis will be important in addressing this question. Moreover, “omics” technologies such as transcriptomics can also resolve the taxonomy of microbes to species and even strain level, yielding more precise information on which bacterial groups are responsible for observed differences in host response to disease. An experimental infection trial incorporating appraisal of shifts in the bacterial community structure using 16S rRNA amplicon techniques (relative abundance) versus changes to bacterial transcription in the same individuals following exposure to a ranavirus would be useful in determining whether bacterial biomass or transcription are more informative in predicting the outcome of infection by a pathogen. A targeted comparison of the metatranscriptomes of wild amphibian populations persisting at sites with endemic ranavirosis versus those from disease-free populations would also help to determine which species of bacteria are transcriptionally linked to ranavirosis under natural conditions.
Microbiome structure vs. stability
The differences between the two studies discussed above could also represent a temporal difference in the skin microbiomes of the animals at the populations sampled. Though a vast number of studies focus on drivers of microbiome structure in the wild, very few focus on microbiome stability and its consequences for the host. Recent experimental work has revealed that the stability of the amphibian skin microbiome is governed by the complexity of the environmental microbial reservoir (Harrison et al. 2019). This gives us strong reason to expect that there is marked variation in the stability of the skin microbiome in wild frogs, which may even vary by population in concert with local environmental conditions. Capture–mark–recapture techniques such as visible implant elastomer tagging or radio-frequency identification tags could be used to examine the temporal stability of the amphibian skin microbiome in the wild by identifying and repeat sampling individual wild amphibians in multi-year frameworks. This would allow for the quantification of the composition, stability, and functional activity of the amphibian skin microbiome and for these traits to be linked to ranaviral disease dynamics throughout longitudinal studies.
Understanding the mechanism underlying microbiome–disease relationships
Whilst present evidence represents an intriguing first demonstration of the potentially important role of the commensal microbiome for determining host response to ranavirosis, the mechanisms and directionality of this relationship are not understood (Campbell et al. 2019; Harrison et al. 2019). Particularly, it is unknown whether the composition of bacteria on amphibian skin is the result of originating from a population with endemic ranavirosis or whether certain bacterial community compositions can predispose a population to suffer outbreaks of ranavirosis. Both ranavirosis (Harrison et al. 2019) and chytridiomycosis (Jani and Briggs 2014) have been shown to perturb the amphibian skin microbiome, which could be expected to generate correlations between microbiome structure and disease status. Understanding the direction of this relationship is critical to appreciate the true potential of using probiotic treatments or bioaugmentation strategies to combat ranaviruses in wild populations. Development of techniques that enable the manipulation of bacterial communities provide the potential to perform experimental infection trials using amphibians with altered cutaneous microbiomes to examine how the presence or absence of key microbial species modulates response to pathogen exposure and subsequent survival. This type of experiment will prove invaluable in disentangling cause and effect in apparent microbiome–disease relationships. This is particularly relevant to when we think about how an individual bacterial species might be interacting with the pathogen. For example, many bacterial species have been shown to demonstrate inhibition of growth of B. dendrobatidis (Antwis and Harrison 2018; Harrison et al. 2020), suggesting a direct interaction. However, microbes can also effect changes to a host’s response to a pathogen indirectly by stimulating host gene expression (Rebollar et al. 2018). To date, no study has discerned whether the microbiome’s role in governing variance in host resistance to ranavirus is driven by direct interaction with the pathogen, or indirect effects on the host. Lack of knowledge regarding this mechanistic pathway remains a clear gap in our understanding that should be addressed as a priority.
Quantifying extrinsic drivers of ranavirosis outbreaks
It is widely appreciated that host–pathogen interactions in many systems are complex and often modulated by external factors, including the environment in which those interactions occur (e.g., James et al. 2015). Recent research conducted within the longitudinally studied R. temporaria populations in the United Kingdom clearly demonstrates that the same is true in the case of ranaviruses and amphibians. The severity of ranavirosis outbreaks within a population has been linked to the use of horticultural chemicals, particularly moluscicides, within or near the water body in which that population resides (North et al. 2015). Likewise, ambient temperature can alter ranavirosis dynamics and infection trajectories (Price et al. 2019), which will have marked implications for variation in disease risk across the geographical distribution of hosts. Here we briefly discuss key knowledge gaps regarding how these abiotic stressors, temperature, and pollutants, influence amphibian disease dynamics, and how they should be addressed.
Host–pollutant–pathogen interactions
The mechanisms by which pollutants may modulate host response to disease are not fully resolved but can broadly be separated into direct and indirect components (Fig. 2). For example, environmental pollutants can act as direct physiological stressors of the amphibian immune system (Robert et al. 2018; Thambirajah et al. 2019), heightening their susceptibility to disease (Fig. 2A). Interestingly, this relationship appears to be reciprocal and the presence of a ranavirus has also been shown to exacerbate the lethality of pesticides (Pochini and Hoverman 2017). Previous studies have shown that herbicides and pesticides can reduce resistance of amphibians to fungal pathogens (e.g., Krynak et al. 2017), perhaps by causing a reduction in anti-microbial peptide (AMP) production (Schadich 2009) and subsequently reduced skin peptide defense against Bd (Davidson et al. 2007; Fig. 2). Both pathways involve a direct effect of the pollutant on the host immune response, which determines subsequent response to a pathogen (Fig. 2A).
Fig. 2.
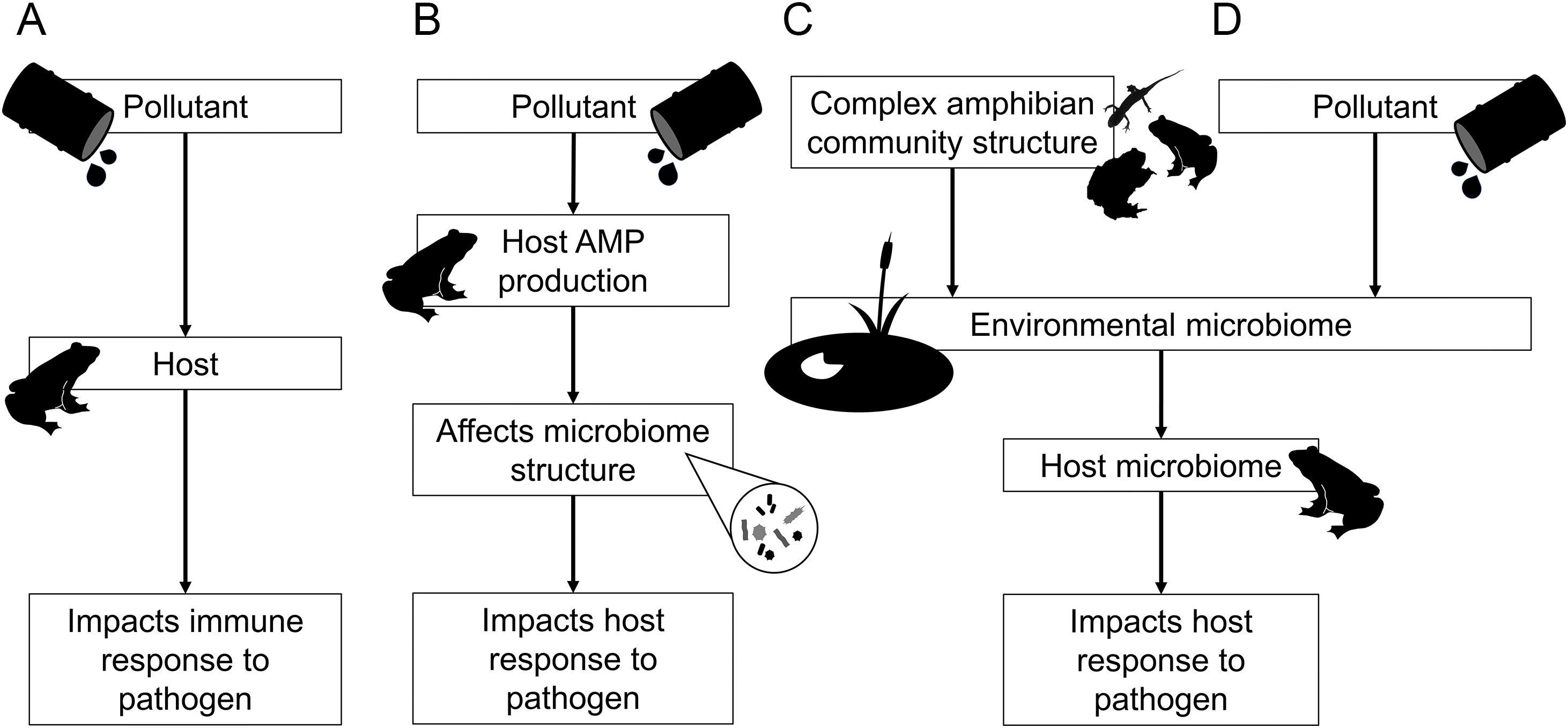
However, despite the important role of the microbiome in governing host response to disease, the potential for pollutants to effect changes in amphibian pathogen resistance indirectly via their influence on the microbiome has received relatively little attention (but see McCoy and Peralta 2018). Both heavy metals from agricultural and industrial run-off, and antimicrobials from pharmaceutical pollutants (Casado et al. 2019), can reduce amphibian gut microbiome diversity (Zhang et al. 2016) and skin microbiome composition (Hernández-Gómez et al. 2020). Pollutant–microbiome relationships may arise either because of a direct effect of the pollutant on the microbiome, or indirectly because pollutants first alter host skin AMP secretion that in turn perturb skin microbial communities (Fig. 2B). Both scenarios represent important pathways for future investigation. Finally, there is strong evidence that the amphibian skin microbiome is populated from environmentally available bacterial species (Kueneman et al. 2014; Longo et al. 2015; Campbell et al. 2019; Harrison et al. 2019). Thus, anything that alters the pool of environmental microbes capable of colonizing the host may also drive differences in subsequent host microbiome structure. Local amphibian community complexity may similarly alter the dynamics and persistence of environmental microbes, for example by introducing novel microbes through immigration (Fig. 2C). Likewise, environmental pollutants can change the diversity of microbial assemblages within both soil and aquatic reservoirs (e.g., Bissett et al. 2013; Muturi et al. 2017; McCoy and Peralta 2018), and indirectly modulate the composition of the amphibian skin microbiome via these environmental effects (Fig. 2D).
Despite growing interest in amphibian microbiome research, studies of host–microbiome–environment interactions remain rare particularly in wild systems (e.g., Varga et al. 2019). Future work should seek to quantify the mechanism by which pollutants alter host susceptibility to disease by simultaneously studying host gene expression and skin microbiome structure in the presence of such contaminants. Surveying at sites with and without historical ranavirus prevalence has already uncovered systematic differences in the structure of the amphibian skin microbiome and the expression of host genes related to the secretion of anti-microbial peptides and inflammation based on disease status (Campbell et al. 2018b, 2019). As amphibians source their skin microbiota from environmental reservoirs, we could expect different geographic locations to influence present amphibian population’s microbiome structure (Campbell et al. 2019; Ross et al. 2019). Therefore, a priority for future studies would be to survey amphibian populations across a wide spatial scale encompassing multiple habitat types, but also varying in ecological gradients of both ranaviral pathogen loads and environmental pollutants. Focal pollutants could include heavy metals near (historically) industrial areas (e.g., Costa et al. 2016), pest-control agents such as molluscicides (North et al. 2015), and fertilizer chemicals (e.g., nitrates) known to affect amphibian physiology and susceptibility to pathogens (e.g., McCoy and Peralta 2018). Developing our understanding of how the amphibian skin microbiome, environmental contaminants, and ranavirosis are inter-linked is important for amphibian conservation efforts and for potential mitigation strategies, such as probiotic treatments; to work in wild systems we must have as full an appreciation as possible about how they will influence and be influenced by the complex interactions between host, pathogen, and environment.
Continued surveillance in the face of environmental change
Ranavirosis outbreaks due to FV3 appear to happen seasonally in the United Kingdom and most incidences of ranavirosis occur when ambient temperatures are 16 °C or higher (Cunningham et al. 1997; Teacher et al. 2010; Price et al. 2019). It has also been experimentally shown that FV3 grows faster at higher temperatures, resulting in a positive correlation between temperature and infection severity in R. temporaria (Price et al. 2019). If current trends in climate warming continue, then the annual time frame during which average monthly temperatures exceed 16 °C will increase and the geographical area over which these conditions are met will spread throughout the United Kingdom (Price et al. 2019). This spread will result in a higher number of populations experiencing ranavirosis outbreaks. It is also likely that climate change will drive distribution shifts of many amphibian species in the United Kingdom (Dunford and Berry 2013), potentially forcing yet more amphibian populations into regions where ranavirosis outbreaks are increasingly likely.
Not only has it been shown that a warming climate will increase the incidence of ranavirosis in the United Kingdom but also that associated climactic instability may be associated with reduced population viability of impacted populations, acting in tandem with age truncation of R. temporaria populations caused by endemic ranavirosis (Campbell et al. 2018a). Age-truncated R. temporaria populations exhibit an over-abundance of younger, smaller breeding individuals (Campbell et al. 2018a). As fecundity of R. temporaria is tightly correlated with body size, per capita recruitment rate of entire populations existing with endemic ranavirosis is lower than that of populations that are disease-free (Campbell et al. 2018a). Population matrix models suggest that this lower recruitment rate potentially heightens the impact of stochastic events that further reduce recruitment such as late frosts, or drought resulting in an increased likelihood of population collapse (Campbell et al. 2018a). As climactic instability also increases due to climate change, the impact of ranavirosis outbreaks on the population level may also be more severe across the ever-expanding range. However, additional population models of R. temporaria have shown that population dynamics are primarily influenced by the survival of adults and not larval or metamorphic individuals (Miaud et al. 1999; Biek et al. 2002). This means the potential for ranavirosis induced population instability may well be unique to the United Kingdom, where ranavirosis impacts adult R. temporaria rather than larval amphibians, which is the case elsewhere in Europe and around the world. Nevertheless, in the United Kingdom, it is important to continue to monitor R. temporaria populations and track the spread of ranavirosis throughout the country as closely as possible.
Little is yet known about the influence of climate on the virulence and spreading potential of CMTV. Understanding the influence of temperature on CMTV through experimental growth assays and in vivo challenge experiments will allow for predictions of how climate change will impact the spread of CMTV throughout continental Europe and the expansion of annual periods of disease outbreaks. The use of e-DNA monitoring tools to quantify environmental loads of ranavirus and subsequent infection risk will be crucial for refining predictive models of outbreak incidence and intensity across environmental gradients, such as intra- and interannual variation in temperature (e.g., Hall et al. 2016; Miaud et al. 2019).
Studies looking into the indirect impacts of CMTV outbreaks on European amphibians are also rare. Field-based studies aimed at understanding how outbreaks of ranavirosis due to CMTV impact the structure of continental European amphibian populations through mechanisms such as age truncation or impinged recruitment will also allow for more accurate predictions of the impact of CMTV on amphibian populations across the continent. Additionally, understanding which regions of continental Europe stand to be more heavily impacted by the spread of ranavirosis requires that we understand which regions harbor the highest number of susceptible species. In vivo challenge experiments could help provide this information. Coupled with an effective surveillance project, likely partially implemented using e-DNA techniques, conducted at the leading edge of disease emergence, precious conservation resources can be targeted towards those regions which are most in peril or those regions where intervention stands the greatest chance of success.
Fill-in ecological blanks for better epidemiological models
Recent events have demonstrated the critical importance of well-parameterised epidemiological models in predicting the spread of emerging diseases and designing and implementing successful mitigation strategies (or not). A small number of studies have attempted to apply various types of epidemiological models to the emergence of ranaviruses in Europe (Campbell et al. 2018a; Duffus et al. 2019), and have provided useful insight into several aspects of host–ranavirus interactions in a European context. Despite this, these studies have also demonstrated that a dearth of knowledge regarding the basic ecology of European amphibians and the ranaviruses that infect them can result in tenuous parameterization of these models from the literature, often drawn from studies focused on distantly related host species or systems with which there is minimal apparent ecological overlap (Campbell et al. 2018a; Duffus et al. 2019). Refining our understanding of host-ranavirus interactions requires that we quantify and incorporate fundamental ecological variation into our models. Within populations, individuals will vary markedly in key life history traits such as growth rate, reproductive output, migration propensity, and (immune) genotype. Likewise, the ranaviral genomic variants hosts are exposed to are expected to vary in replication rates and subsequent transmission potential. Collectively these traits will interact to influence parameters such as the likelihood, frequency, and intensity of ranavirus infection within, and transmission among, amphibian populations. Tackling these knowledge gaps will undoubtedly require long term data sets of marked individuals paired with frequent monitoring, quantification, and genotyping of ranaviral genomic variants to understand the factors governing long term probability of persistence of both pathogen and hosts in natural populations. Using these long-term data sets to quantify rates of disease transmission within and among populations will undoubtedly lead to strong epidemiological models, increasing the accuracy with which researchers can make predictions about the spread of these deadly emerging pathogens.
Conclusion
Ranaviruses are an emerging disease threat to European amphibian populations. Both directly, through elevated mortality, and indirectly, through the effects of endemic disease on host populations, ranaviruses have been found to cause catastrophic and sustained declines in amphibians in the United Kingdom and on continental Europe. As climate change drives global temperature increases and climatic instability, the pace of ranaviral emergence and the severity of ranavirosis outbreaks in Europe are likely to follow suit. This will result in the exposure of more populations and additional species over a wider annual time period and a reduction in the long-term viability of impacted populations. In light of this, it is imperative that we continue to monitor the spread of ranaviruses throughout Europe and to assess which currently naïve European amphibian species are likely to be most vulnerable to infection, allowing for the targeting of conservation efforts in regions where the impact of ranaviral emergence is likely to be particularly severe.
Happily, recent research suggests that the amphibian skin microbiome may offer a source of potential mitigation strategies in the form of probiotic treatment and environmental bioaugmentation, as it has done in response to the chytrid fungi (Kueneman et al. 2016; Antwis and Harrison 2018; Harrison et al. 2020). For the potential efficacy of such strategies to be appraised, a number of fundamental research questions need to first be addressed. Primarily, we must aim to assess the generality of relationships between the amphibian skin microbiome and ranavirosis in a wide array of systems. Additional questions regarding the measure by which we judge the importance of a microbial species in a microbiome and the subsequent impact of a complex environment on host/microbiome–pathogen interactions will need to be addressed. European research has been critical in generating knowledge regarding ranaviruses and their impact on amphibians. The proximity within which many European amphibian populations live to humans, coupled with the proven potential for significant public engagement in the study of ranavirosis in Europe, presents unique opportunities to address these important research questions that could lead to advances in amphibian conservation not just in Europe but around the world.
References
Allain SJR, and Duffus ALJ. 2019. Emerging infectious disease threats to European herpetofauna. The Herpetological Journal, 29: 189–206.
Aly SM, Abdel-Galil Ahmed Y, Abdel-Aziz Ghareeb A, and Mohamed MF. 2008. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish & Shellfish Immunology, 25: 128–136.
Antwis RE, and Harrison XA. 2018. Probiotic consortia are not uniformly effective against different amphibian chytrid pathogen isolates. Molecular Ecology, 27: 577–589.
Ariel E, Kielgast J, Svart HE, Larsen K, Tapiovaara H, Jensen BB, et al. 2009. Ranavirus in wild edible frogs Pelophylax kl. esculentus in Denmark. Diseases of Aquatic Organisms, 85: 7–14.
Ariel E, Holopainen R, Olesen NJ, and Tapiovaara H. 2010. Comparative study of ranavirus isolates from cod (Gadus morhua) and turbot (Psetta maxima) with reference to other ranaviruses. Archives of Virology, 155: 1261–1271.
Balseiro A, Dalton KP, Del Cerro A, Marquez I, Cunningham AA, Parra F, et al. 2009. Pathology, isolation and molecular characterisation of a ranavirus from the common midwife toad Alytes obstetricans on the Iberian Peninsula. Diseases of Aquatic Organisms, 84: 95–104.
Bates KA, Shelton JMG, Mercier VL, Hopkins KP, Harrison XA, Petrovan SO, et al. 2019. Captivity and infection by the fungal pathogen Batrachochytrium salamandrivorans perturb the amphibian skin microbiome. Frontiers in Microbiology, 10: 1834.
Becker MH, Walke JB, Cikanek S, Savage AE, Mattheus N, Santiago CN, et al. 2015. Composition of symbiotic bacteria predicts survival in Panamanian golden frogs infected with a lethal fungus. Proceedings of the Royal Society B: Biological Sciences, 282: 20142881.
Biek R, Funk WC, Maxell BA, and Mills LS. 2002. What is missing from insights amphibian decline ecological sensitivity analysis. Conservation Biology, 16: 728–734.
Bissett A, Brown MV, Siciliano SD, and Thrall PH. 2013. Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecology Letters, 16: 128–139.
Campbell LJ, Garner TWJ, Tessa G, Scheele BC, Griffiths AGF, Wilfert L, et al. 2018a. An emerging viral pathogen truncates population age structure in a European amphibian and may reduce population viability. PeerJ, 6: e5949.
Campbell LJ, Hammond SA, Price SJ, Sharma MD, Garner TWJ, Birol I, et al. 2018b. A novel approach to wildlife transcriptomics provides evidence of disease-mediated differential expression and changes to the microbiome of amphibian populations. Molecular Ecology, 27: 1413–1427.
Campbell LJ, Garner TWJ, Hopkins K, Griffiths AGF, and Harrison XA. 2019. Outbreaks of an emerging viral disease covary with differences in the composition of the skin microbiome of a wild United Kingdom amphibian. Frontiers in Microbiology, 10: 1245.
Casado J, Brigden K, Santillo D, and Johnston P. 2019. Screening of pesticides and veterinary drugs in small streams in the European Union by liquid chromatography high resolution mass spectrometry. Science of the Total Environment, 670: 1204–1225.
Chinchar VG, Hick P, Ince IA, Jancovich JK, Marschang R, Qin Q, et al. 2017. ICTV virus taxonomy profile: Iridoviridae. Journal of General Virology, 98: 890–891.
Claytor SC, Subramaniam K, Landrau-Giovannetti N, Chinchar VG, Gray MJ, Miller DL, et al. 2017. Ranavirus phylogenomics: signatures of recombination and inversions among bullfrog ranaculture isolates. Virology, 511: 330–343.
Costa S, Lopes I, Proen¸a DN, Ribeiro R, and Morais PV. 2016. Diversity of cutaneous microbiome of Pelophylax perezi populations inhabiting different environments. Science of the Total Environment, 572: 995–1004.
Crilly R, and Esteban A. 2013. Small versus large-scale, multi-fleet fisheries: the case for economic, social and environmental access criteria in European fisheries. Marine Policy, 37: 20–27.
Cunningham AA, Langton TE, Bennett PM, Lewin JF, Drury SE, Gough RE, et al. 1997. Pathological and microbiological findings from incidents of unusual mortality of the common frog (Rana temporaria). Philosophical Transactions of the Royal Society B: Biological Sciences, 351: 1539–1557.
Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, and Speare R. 1999. Emerging infectious diseases and amphibian population declines. Emerging Infectious Diseases, 5: 735–748.
Davidson C, Benard MF, Shaffer HB, Parker JM, O’Leary C, Conlon JM, et al. 2007. Effects of chytrid and carbaryl exposure on survival, growth and skin peptide defenses in foothill yellow-legged frogs. Environmental Science & Technology, 41: 1771–1776.
Duffus ALJ, Nichols RA, and Garner TWJ. 2013. Investigations into the life history stages of the common frog (Rana temporaria) affected by an amphibian ranavirus in the United Kingdom. Herpetological Review, 44: 260–263.
Duffus ALJ, Garner TWJ, Davis AR, Dean AW, and Nichols RA. 2017. Phylogenetic analysis of 24 ranavirus isolates from English amphibians using 2 partial loci. Journal of Emerging Diseases and Virology, 3: 1–10.
Duffus ALJ, Garner TWJ, Nichols RA, Standridge JP, and Earl JE. 2019. Modelling ranavirus transmission in populations of common frogs (Rana temporaria) in the United Kingdom. Viruses, 11: 556.
Dunford RW, and Berry PM. 2013. Climate change modelling of English amphibians and reptiles: report to Amphibian and Reptile Conservation Trust (ARC-Trust). ARC-Trust. 43 p.
Earl JE, and Gray MJ. 2014. Introduction of ranavirus to isolated wood frog populations could cause local extinction. EcoHealth, 11: 581–592.
Fuller RA, and Gaston KJ. 2009. The scaling of green space coverage in European cities. Biology Letters, 5: 352–355.
Hall EM, Crespi EJ, Goldberg CS, and Brunner JL. 2016. Evaluating environmental DNA-based quantification of ranavirus infection in wood frog populations. Molecular Ecology Resources, 16(2): 423–433.
Harris RN, James TY, Lauer A, Simon MA, and Patel A. 2006. Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. EcoHealth, 3: 53–56.
Harris RN, Brucker R, Walke J, Becker M, and Schwantes C. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. The ISME Journal, 3: 818–824.
Harrison XA, and Cameron SJ. 2020. Analytical approaches for microbiome research. In Microbiomes of plants, soils and animals: an integrated approach. Edited by R Antwis, X Harrison, and M Cox. Cambridge Unversity Press, Cambridge, UK. pp. 8–28.
Harrison XA, Price SJ, Hopkins K, Leung WTM, Sergeant C, and Garner TWJ. 2019. Diversity-stability dynamics of the amphibian skin microbiome and susceptibility to a lethal viral pathogen. Frontiers in Microbiology, 10: 2883.
Harrison XA, Sewell T, Fisher M, and Antwis RE. 2020. Designing probiotic therapies with broad-spectrum activity against a wildlife pathogen. Frontiers in Microbiology, 10: 3134.
Hernández-Gómez O, Wuerthner V, and Hua J. 2020. Amphibian host and skin microbiota response to a common agricultural antimicrobial and internal parasite. Microbial Ecology, 79: 175–191.
Hyatt AD, Gould AR, Zupanovic Z, Cunningham AA, Hengstberger SG, Whittington RJ, et al. 2000. Comparative studies of piscine and amphibian iridoviruses. Archives of Virology, 145: 301–331.
James TY, Toledo LF, Rödder D, da Silva Leite D, Belasen AM, Betancourt-Román CM, et al. 2015. Disentangling host, pathogen, and environmental determinants of a recently emerged wildlife disease: lessons from the first 15 years of amphibian chytridiomycosis research. Ecology and Evolution, 5: 4079–4097.
Jani AJ, and Briggs CJ. 2014. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proceedings of the National Academy of Sciences of the United States of America, 111: E5049–E5058.
Jensen NJ, Bloch B, and Larsen JL. 1979. The ulcus-syndrome in cod (Gadus morhua). III. A preliminary virological report. Nordisk Veterinaermedicin, 31: 436–442.
Krynak KL, Burke DJ, and Benard MF. 2017. Rodeo™ herbicide negatively affects Blanchard’s Cricket Frogs (Acris blanchardi) survival and alters the skin-associated bacterial community. Journal of Herpetology, 51: 402–410.
Kueneman JG, Parfrey LW, Woodhams DC, Archer HM, Knight R, and McKenzie VJ. 2014. The amphibian skin-associated microbiome across species, space and life history stages. Molecular Ecology, 23: 1238–1250.
Kueneman JG, Woodhams DC, Harris R, Archer HM, Knight R, and McKenzie VJ. 2016. Probiotic treatment restores protection against lethal fungal infection lost during amphibian captivity. Proceedings of the Royal Society B: Biological Sciences, 283: 20161553.
Lawson B, Petrovan SO, and Cunningham AA. 2015. Citizen science and wildlife disease surveillance. EcoHealth, 12: 693–702.
Liu CH, Chiu CH, Wang SW, and Cheng W. 2012. Dietary administration of the probiotic, Bacillus subtilis E20, enhances the growth, innate immune responses, and disease resistance of the grouper, Epinephelus coioides. Fish & Shellfish Immunology, 33: 699–706.
Longo AV, Savage AE, Hewson I, and Zamudio KR. 2015. Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. Royal Society Open Science, 2: 140377.
Marschang RE. 2011. Viruses infecting reptiles. Viruses, 3: 2087–2126.
McCoy KA, and Peralta AL. 2018. Pesticides could alter amphibian skin microbiomes and the effects of Batrachochytrium dendrobatidis. Frontiers in Microbiology, 9: 748.
Miaud C, Guyétant R, and Elmberg J. 1999. Variations in life-history traits in the common frog Rana temporaria (Amphibia: Anura): a literature review and new data from the French Alps. Journal of Zoology, 249: 61–73.
Miaud C, Pozet F, Gaudin NCG, Martel A, Pasmans F, and Labrut S. 2016. Ranavirus causes mass die-offs of alpine amphibians in the southwestern Alps, France. Journal of Wildlife Diseases, 52: 242–252.
Miaud C, Arnal V, Poulain M, Valentini A, and Dejean T. 2019. eDNA increases the detectability of ranavirus infection in an alpine amphibian population. Viruses, 11: 526.
Muturi EJ, Donthu RK, Fields CJ, Moise IK, and Kim CH. 2017. Effect of pesticides on microbial communities in container aquatic habitats. Scientific Reports, 7: 44565.
North AC, Hodgson DJ, Price SJ, and Griffiths AGF. 2015. Anthropogenic and ecological drivers of amphibian disease (ranavirosis). PLoS ONE, 10: e0127037.
Pochini KM, and Hoverman JT. 2017. Reciprocal effects of pesticides and pathogens on amphibian hosts: the importance of exposure order and timing. Environmental Pollution, 221: 359–366.
Price SJ. 2015. Comparative genomics of amphibian-like ranaviruses, nucleocytoplasmic large DNA viruses of poikilotherms. Evolutionary Bioinformatics, 11(Suppl. 2): 71–82.
Price SJ, Garner TWJ, Nichols RA, Balloux F, Ayres C, Mora-Cabello de Alba A, et al. 2014. Collapse of amphibian communities due to an introduced Ranavirus. Current Biology, 24: 2586–2591.
Price SJ, Garner T, Cunningham A, Langton T, and Nichols R. 2016. Reconstructing the emergence of a lethal infectious disease of wildlife supports a key role for spread through translocations by humans. Proceedings of the Royal Society B: Biological Sciences, 283: 20160952.
Price SJ, Cunningham AA, Price SJ, Wadia A, Wright ON, and Leung WTM. 2017a. Screening of a long-term sample set reveals two Ranavirus lineages in British herpetofauna. PLoS ONE, 12: e0184768.
Price SJ, Ariel E, Maclaine A, Rosa GM, Gray MJ, Brunner JL, et al. 2017b. From fish to frogs and beyond: impact and host range of emergent ranaviruses. Virology, 511: 272–279.
Price SJ, Leung WTM, Owen CJ, Puschendorf R, Sergeant C, Cunningham AA, et al. 2019. Effects of historic and projected climate change on the range and impacts of an emerging wildlife disease. Global Change Biology, 25: 2648–2660.
Ran C, Carrias A, Williams MA, Capps N, Dan BCT, Newton JC, et al. 2012. Identification of Bacillus strains for biological control of catfish pathogens. PLoS ONE, 7: e45793.
Rebollar EA, Antwis RE, Becker MH, Belden LK, Bletz MC, Brucker RM, et al. 2016. Using “omics” and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Frontiers in Microbiology, 7: 68.
Rebollar EA, Gutiérrez-Preciado A, Noecker C, Eng A, Hughey MC, Medina D, et al. 2018. The skin microbiome of the neotropical frog Craugastor fitzingeri: inferring potential bacterial-host-pathogen interactions from metagenomic data. Frontiers in Microbiology, 9: 466.
Rijks JM, Saucedo B, Spitzen-van der Sluijs A, Wilkie GS, van Asten AJAM, van den Broek J, et al. 2016. Investigation of amphibian mortality events in wildlife reveals an on-going ranavirus epidemic in the north of the Netherlands. PLoS ONE, 11: e0157473.
Robert J, McGuire CC, Kim F, Nagel SC, Price SJ, Lawrence BP, et al. 2018. Water contaminants associated with unconventional oil and gas extraction cause immunotoxicity to amphibian tadpoles. Toxicological Sciences, 166: 39–50.
Rosa GM, Sabino-Pinto J, Laurentino TG, Martel A, Pasmans F, Rebelo R, et al. 2017. Impact of asynchronous emergence of two lethal pathogens on amphibian assemblages. Scientific Reports, 7: 43260.
Ross AA, Rodrigues Hoffmann A, and Neufeld JD. 2019. The skin microbiome of vertebrates. Microbiome, 7: 79.
Saucedo B, Hughes J, Spitzen-van der Sluijs A, Kruithof N, Schills M, Rijks JM, et al. 2018. Ranavirus genotypes in the Netherlands and their potential association with virulence in water frogs (Pelophylax spp.). Emerging Microbes & Infections, 7: 56.
Saucedo B, Garner TWJ, Kruithof N, Allain SJR, Goodman MJ, Cranfield RJ, et al. 2019. Common midwife toad ranaviruses replicate first in the oral cavity of smooth newts (Lissotriton vulgaris) and show distinct strain-associated pathogenicity. Scientific Reports, 9: 4453.
Schadich E. 2009. Skin peptide activities against opportunistic bacterial pathogens of the African Clawed Frog (Xenopus laevis) and three Litoria frogs. Journal of Herpetology, 43: 173–183.
Stöhr AC, López-Bueno A, Blahak S, Caeiro MF, Rosa GM, Alves de Matos AP, et al. 2015. Phylogeny and differentiation of reptilian and amphibian ranaviruses detected in Europe. PLoS ONE, 10: e0118633.
Teacher AGF, Garner TWJ, and Nichols RA. 2009a. Evidence for directional selection at a novel major histocompatibility class I marker in wild common frogs (Rana temporaria) exposed to a viral pathogen (Ranavirus). PLoS ONE, 4: e4616.
Teacher AGF, Garner TWJ, and Nichols RA. 2009b. Population genetic patterns suggest a behavioural change in wild common frogs (Rana temporaria) following disease outbreaks (Ranavirus). Molecular Ecology, 18: 3163–3172.
Teacher AGF, Cunningham AA, and Garner TWJ. 2010. Assessing the long-term impact of Ranavirus infection in wild common frog populations. Animal Conservation, 13: 514–522.
Thambirajah AA, Koide EM, Imbery JJ, and Helbing CC. 2019. Contaminant and environmental influences on thyroid hormone action in amphibian metamorphosis. Frontiers in Endocrinology, 10: 276.
Varga JFA, Bui-Marinos MP, and Katzenback BA. 2019. Frog skin innate immune defences: sensing and surviving pathogens. Frontiers in Immunology, 9: 3128.
Warne RW, Kirschman L, and Zeglin L. 2019. Manipulation of gut microbiota during critical developmental windows affects host physiological performance and disease susceptibility across ontogeny. Journal of Animal Ecology, 88: 845–856.
Zhang W, Guo R, Yang Y, Ding J, and Zhang Y. 2016. Long-term effect of heavy-metal pollution on diversity of gastrointestinal microbial community of Bufo raddei. Toxicology Letters, 258: 192–197.
Information & Authors
Information
Published In

FACETS
Volume 5 • Number 1 • January 2020
Pages: 598 - 614
Editor: David Lesbarrères
History
Received: 18 February 2020
Accepted: 11 May 2020
Version of record online: 6 August 2020
Notes
This paper is part of a Collection titled “Ranavirus research: 10 years of global collaboration”.
Copyright
© 2020 Campbell et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
LJC and XAH conceived and designed the study.
All drafted or revised the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Lewis J. Campbell, Alice H. Pawlik, and Xavier A. Harrison. 2020. Amphibian ranaviruses in Europe: important directions for future research. FACETS.
5(1): 598-614. https://doi.org/10.1139/facets-2020-0007
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Gene functions of the Ambystoma altamirani skin microbiome vary across space and time but potential antifungal genes are widespread and prevalent
2. Ontogeny drives shifts in skin bacterial communities in facultatively paedomorphic salamanders
3. Diet- and salinity-induced modifications of the gut microbiota are associated with differential physiological responses to ranavirus infection in
Rana sylvatica
4. Infectious disease threats to amphibians in Greece: new localities positive for Batrachochytrium dendrobatidis
5. Recent insights into aquatic viruses: Emerging and reemerging pathogens, molecular features, biological effects, and novel investigative approaches
