Phylogenomic characterization of ranaviruses isolated from cultured fish and amphibians in Thailand
Abstract
Ranaviruses are emerging pathogens associated with worldwide epizootics in farmed and wild ectothermic vertebrates. In this study, we determined the full genomes of eight ranaviruses isolated from marbled sleeper goby (Oxyeleotris marmorata), goldfish (Carassius auratus), guppy (Poecilia reticulata), tiger frog (Hoplobatrachus tigerinus), Asian grass frog (Fejervarya limnocharis), and East Asian bullfrog (H. rugulosus) cultured or imported into Thailand. These ranaviral isolates induced the same cytopathic effects (i.e., progression of coalescing round plaques) in epithelioma papulosum cyprini (EPC) cell cultures. Transmission electron microscopy of infected EPC cells revealed cytoplasmic viral particles with ultrastructural features typical for ranaviruses. Pairwise genetic comparisons of the complete major capsid protein coding sequences from the Thai ranaviruses displayed the highest identity (99.8%–100%) to a ranavirus (tiger frog virus; TFV) isolated from diseased tiger frogs cultured in China, a slightly lower identity (99.3%–99.4%) to a ranavirus (Wamena virus; WV) isolated from diseased green tree pythons (Morelia viridis) illegally exported from Papua New Guinea, and a lower identity to 35 other ranaviruses (93.7%–98.6%). Phylogenomic analyses supported the eight Thai ranaviruses, Chinese TFV, and WV as a subclade within a larger frog virus 3 clade. Our findings confirm the spread of TFV among cultured fish and amphibians in Asia and likely in reptiles in Oceania. Biosecurity measures are needed to ensure TFV does not continue to spread throughout Southeast Asia and to other parts of the world via international trade.
Introduction
Globally, aquaculture is an essential food-producing sector and it has played a crucial role in the Thai economy for decades (Food and Agriculture Organization of the United Nations 2020). Between 1998 and 2017, Thailand’s average annual aquaculture production was approximately 1.1 million tons per year, with freshwater aquaculture production contributing nearly half of the annual production (Department of Fisheries Thailand 2019a). Of the total Thai freshwater aquaculture production in 2017, 4000 tons were contributed from ranaculture products including live frogs, chilled and frozen frog legs, and whole frogs (Department of Fisheries Thailand 2019b). However, diseases including those caused by ranaviruses have emerged as an impediment to the continued growth of ranaculture around the world including Asia (Zhang et al. 2001; Weng et al. 2002), North America (Miller et al. 2007), and South America (Mazzoni et al. 2009).
Members of the genus Ranavirus (family Iridoviridae; subfamily Alphairidovirinae) possess an enveloped nucleocapsid (150–200 nm in diameter) that encloses a double-stranded DNA genome (103–220 kbp) (Chinchar et al. 2017). The genus includes the following species recognized by the International Committee on Taxonomy of Viruses: Frog virus 3 (FV3), Ambystoma tigrinum virus, Common midwife toad virus, Epizootic haematopoietic necrosis virus (EHNV), Santee-Cooper ranavirus, and Singapore grouper iridovirus (International Committee on Taxonomy of Viruses 2020). Ranaviruses are emerging pathogens infecting at least 175 species and 52 families of ectothermic vertebrates (fish, amphibians, and reptiles) across six continents (Duffus et al. 2015). FV3 has the widest-known host and geographic range of all ranaviruses and includes Asian strains isolated from diseased fish, amphibians, and reptiles reared for food in China and Thailand. FV3 strains have negatively impacted the culture of tiger frogs (Hoplobatrachus tigerinus) and East Asian bullfrogs (H. rugulosus) in Thailand and China (Kanchanakhan et al. 1998, 1999; Weng et al. 2002), Chinese softshell turtles (Pelodiscus sinensis) in China (Chen et al. 1999), pig frogs (Lithobates grylio) in China (Zhang et al. 2001), and marbled sleeper goby (Oxyeleotris marmorata) in Thailand (Prasankok et al. 2005). Ranaviruses have been identified as a global threat to free-ranging amphibian populations as they have the potential to cause population declines and extinctions (Duffus and Cunningham 2010; Teacher et al. 2010; Miller et al. 2011; Earl and Gray 2014; Price et al. 2014; Peace et al. 2019). Ranaviral infections in amphibians and EHNV infection in rainbow trout (Oncorhynchus mykiss) and redfin perch (Perca fluviatilis) are notifiable to the World Organization for Animal Health (2019).
Over the last two decades, strains of FV3 have repeatedly been isolated during mortality events in ranaculture and aquaculture facilities in Thailand. From 1998 to 2002, ranaculture facilities in nine provinces in the central part of Thailand experienced moderate (20%–50%) to high (95%) mortalities in cultured tadpoles and juvenile and adult tiger frogs (Kanchanakhan 1998, 2011; Kanchanakhan et al. 1999). Diseased frogs displayed ulcerative lesions on the rostrum (lips and mouth) and dorsal part of the body and legs. Bacteria were not isolated from the liver, kidney, and spleen in the early stages of the disease; however, bacteria were cultured from frogs that experienced chronic high mortality infections. Skin and internal tissue homogenates from diseased frogs and tadpoles were processed for viral isolation in epithelioma papulosum cyprini (EPC) cells and resulted in the recovery of passageable agents in 65% (70/107) of the individuals tested from 1998 to 2002 (Kanchanakhan 2011). Physicochemical and ultrastructural features of the isolates supported them as members of the family Iridoviridae. In 2004, high mortality (>50%) in Asian grass frogs (Fejervarya limnocharis) was reported on a private fish farm in Northeast Thailand following importation from Cambodia (Kanchanakhan et al. 2004). The diseased frogs showed skin ulcerations on the rostrum and legs. A replicating agent was isolated in EPC cultures and determined to be an iridovirus based on its physicochemical and ultrastructural features. PCR amplification and sequencing of the iridoviral partial major capsid protein (MCP) and ATPase genes demonstrated it was nearly identical (98%–99%) to a strain of FV3 known as tiger frog virus (TFV), previously isolated from diseased tiger frog tadpoles cultured in China in 2000 (Weng et al. 2002) and hereafter referred to as Chinese TFV. In 2000, high mortalities in cultured marbled sleeper goby were reported in the Nakonpathom province of Central Thailand (Prasankok et al. 2005). Fish displayed ulcerative lesions on the body and around the mouth. No consistent parasites or bacteria were observed or cultured from the diseased fish. A replicating agent was isolated in EPC cultures and again determined to be an iridovirus based on its physicochemical and ultrastructural features. PCR amplification and sequencing of the iridoviral partial MCP gene demonstrated the isolate was nearly identical (98%–99%) to the aforementioned Chinese TFV and Thai ranaviruses isolated from imported Asian grass frogs and cultured tiger frogs.
Herein, we describe the in vitro growth (i.e., observed cytopathic effects), ultrastructural, and phylogenomic characteristics of ranaviruses isolated from fish and amphibians cultured in Thailand for food or ornamental purposes. The determined complete genome sequences are the first to be publicly available for ranaviruses isolated from Thai ectothermic vertebrates. This genomic data was used in phylogenomic analyses that revealed TFV, a strain of FV3, has spread among facilities in Asia rearing fish and amphibians, and perhaps also in free-ranging reptiles in Oceania.
Materials and methods
Clinical history
From 1998 to 2017, a series of ranaviruses was isolated from diseased fish, tadpoles, and frogs submitted by Thai aquaculture and ranaculture facilities to the Aquatic Animal Health Research and Development Division (AAHRDD), Department of Fisheries, Bangkok, Thailand. Details of AAHRDD diagnostic evaluations (e.g., bacteriology, virology, transmission electron microscopy, and conventional PCR) that resulted in ranaviral isolates from tiger frogs (isolate AV9803) in 1998, marbled sleeper goby in 2000 (isolate D2008) (Fig. 1A), and imported Asian grass frogs in 2004 (isolate D03-034) (Fig. 1B) have previously appeared (Kanchanakhan 1998, 2011; Kanchanakhan et al. 1999, 2004; Prasankok et al. 2005). Ranaviruses isolated by AAHRDD during a health assessment of overtly healthy guppies (Poecilia reticulata) in 2001 (isolate F2112) and goldfish (Carassius auratus) in 2002 (isolate F0207) were also included in this study (S. Kanchanakhan, personal observation, 2001, 2002). In 2011 and 2016, Thai facilities rearing adult East Asian bullfrogs reported elevated mortality with moribund frogs displaying ulcerative lesions on the dorsal part of the body and legs (Figs. 1C and 1D). In 2017, a Thai facility rearing East Asian bullfrog tadpoles reported elevated mortality with moribund tadpoles displaying cutaneous ulcerations and edema (J. Polchana, personal observation, 2017). Samples from these East Asian bullfrog tadpoles and frogs were submitted to AAHRDD for virology and resulted in three additional ranaviral isolates (D11-067, VD16-006, and VD17-007) used in this study. The Thai ranaviral isolates analyzed in this study and their respective host and temporospatial details are provided in Table 1.
Fig. 1.
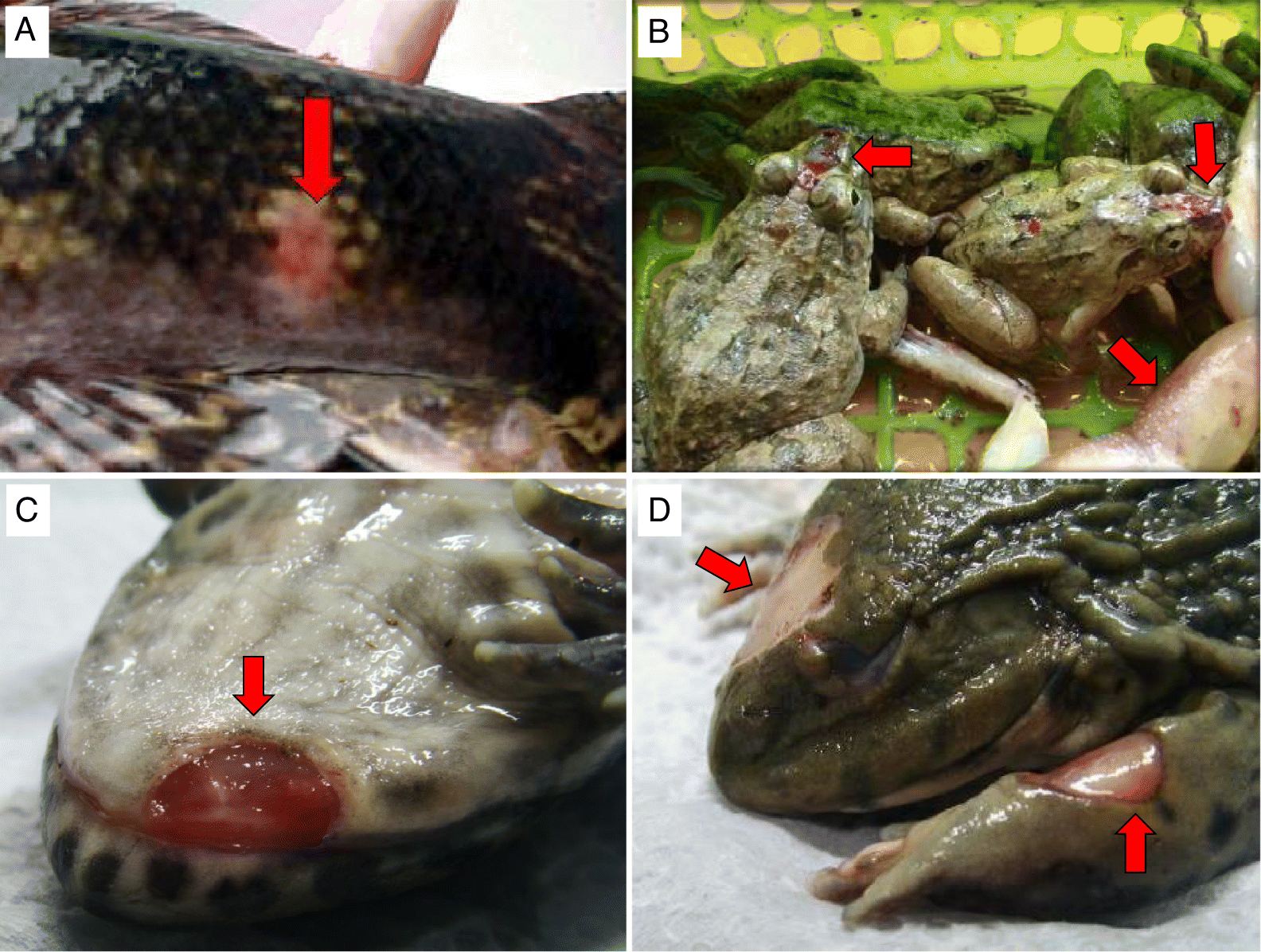
Table 1.
| Viral name (abbreviation) | Isolate | Host | Naked virion apex–apex (nm), mean (SD) | Naked virion side-side (nm), mean (SD) | Enveloped virion apex–apex (nm), mean (SD) | Enveloped virion side–side (nm), mean (SD) |
|---|---|---|---|---|---|---|
| Tiger frog virus (TFV-1998) | AV9803 | Tiger frog (Hoplobatrachus tigerinus) | 157.3 (2.1) | 132.5 (3.2) | 196.5 (4.9) | 187.3 (5.7) |
Oxyeleotris marmorata ranavirus (OMRV) | D2008 | Marbled sleeper goby (Oxyeleotris marmorata) | 158.8 (1.2) | 127.6 (1.6) | 201.3 (2.8) | 186.3 (7.6) |
Poecilia reticulata ranavirus (PPRV) | F2112 | Guppy (Poecilia reticulata) | 158.2 (1.6) | 123.5 (2.3) | 203.7 (4.0) | 187.3 (3.7) |
| Goldfish ranavirus (GFRV) | F0207 | Goldfish (Carassius auratus) | 150.2 (1.2) | 125.5 (1.6) | 185.5 (1.5) | 164.3 (3.1) |
Asian grass frog ranavirus (AGFRV) | D03-034 | Asian grass frog (Fejervarya limnocharis) | 158.7 (1.0) | 128.9 (1.4) | 201.7 (2.1) | 185.3 (3.8) |
East Asian bullfrog ranavirus (EABRV-2011) | D11-067 | East Asian bullfrog (H. rugulosus) | 158.8 (1.3) | 130.4 (1.6) | 194.9 (4.5) | 177.6 (5.6) |
East Asian bullfrog ranavirus (EABRV-2016) | VD-16-006 | East Asian bullfrog (H. rugulosus) | 158.9 (1.7) | 129.8 (2.7) | 206.0 (3.3) | 185.8 (4.0) |
East Asian bullfrog ranavirus (EABRV-2017) | VD-17-007 | East Asian bullfrog (H. rugulosus) | NO | NO | NO | NO |
Note
Means (±standard deviation, SD) are based on the measurement of 20 unenveloped virions and 3–16 enveloped virions per isolate. NO, not observed.
In vitro propagation of ranaviruses
Eight presumed ranaviral isolates recovered at AAHRDD from cultured frogs and fish from 1998 through 2017 were shipped to the University of Florida Wildlife and Aquatic Veterinary Disease Laboratory (WAVDL), Gainesville, Florida, USA (Table 1). At WAVDL, 1 mL of each isolate was inoculated onto a confluent monolayer of EPC cells within 75 cm2 flasks maintained in modified Eagle’s medium with 10% fetal bovine serum. Inoculated EPC cultures were maintained at 25 °C and observed daily for cytopathic effects (CPE).
Transmission electron microscopy (TEM)
Seven of the isolates (VD-17-007 was not submitted for TEM) were individually propagated in 75 cm2 flasks of EPC cells until CPE was observed. The cells were fixed in 15 mL of modified Karnovsky’s fixative (2P + 2G, 2% formaldehyde prepared from paraformaldehyde and 2% glutaraldehyde in 0.1 mol/L cacodylate buffer pH 7.4) at room temperature for 1 h. Then the monolayer was washed in cacodylate buffer, scraped off the flask, and pelleted (3000g at 4 °C for 10 min). The pellet was shipped in phosphate-buffered saline overnight on ice packs to the University of Texas Medical Branch Department of Pathology Electron Microscopy Laboratory (UTMB-EML). At UTMB-EML, the cell pellet was washed in cacodylate buffer and left in 2P + 2G fixative overnight at 4 °C. The next day the cell pellet was washed twice in cacodylate buffer, postfixed in 1% OsO4 in 0.1 mol/L cacodylate buffer pH 7.4, en bloc stained with 2% aqueous uranyl acetate, dehydrated in ascending concentrations of ethanol, processed through propylene oxide, and embedded in Poly/Bed 812 epoxy plastic (Polysciences). Ultrathin sections were cut on a Leica ULTRACUT EM UC7 ultramicrotome (Leica Microsystems), stained with 0.4% lead citrate, and examined in a JEM-1400 electron microscope (JEOL USA) at 80 kV.
DNA extraction, whole genome sequencing and assembly
Each viral isolate was inoculated into two 75 cm2 flasks of EPC cells and harvested when extensive CPE was observed (48–72 h postinfection). Then, the cells were subjected to three rounds of freezing–thawing prior to clarification of the supernatant by centrifugation at 5520g for 20 min at 4 °C. The clarified supernatant was then centrifuged at 100 000g for 1.15 h at 4 °C to obtain a viral pellet. Pelleted virus was then resuspended in 360 μL of animal tissue lysis (ATL) buffer prior to extraction of viral genomic DNA using a DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instruction. DNA sequencing libraries were generated using a TruSeq DNA PCR-Free Library Preparation kit (Illumina) and sequencing was performed using a v3 chemistry 600 cycle kit on a MiSeq sequencer (Illumina). De novo assembly of the paired-end reads was performed in SPAdes 3.5.0 (Bankevich et al. 2012). The quality of the genome assembly was verified by mapping the reads back to the consensus sequence in Bowtie2 2.1.0 (Langmead and Salzberg 2012) and visually inspecting the alignment in Tablet 1.14.10.20 (Milne et al. 2013).
Genome annotation and phylogenomic analyses
The genomes of the Thai ranaviruses were annotated using the Genome Annotation Transfer Utility (Tcherepanov et al. 2006) with TFV (GenBank Accession No. AF389451.1) used as the reference genome. Putative open reading frames (ORFs) were added or removed by comparison to the FV3 genome annotation (Tan et al. 2004) and using GenemarkS (Besemer et al. 2001) and CLC Genomics Workbench 12.0 based on the following criteria: (i) only ORFs larger than 120 nucleotides were considered; (ii) ORFs were not allowed to overlap with neighboring ORFs by more than 25%; and (iii) in the event of overlapping, only the larger ORF was annotated or overlapping ORFs were permitted provided they represent existing ranaviral orthologs (Bennett et al. 2017; Koda et al. 2018). Gene function was predicted based on BLASTP searches against the National Center for Biotechnology Information GenBank nonredundant protein sequence database. The eight Thai ranaviral genomes and 36 fully sequenced ranaviral genomes retrieved from GenBank (Table 2) were aligned using Mauve 2.4 software (Darling et al. 2004) to visualize genomic inversions and obtain the locally collinear block (LCB) alignments. The LCB alignments were then concatenated in Geneious v.10.0.2 (Kearse et al. 2012) and a Maximum Likelihood (ML) analysis was performed in IQ-TREE (http://iqtree.cibiv.univie.ac.at/) with the Bayesian information criterion used to determine the best model fit and 1000 nonparametric bootstraps to determine node support (Nguyen et al. 2015). To elucidate the relationship of Thai ranaviruses to the Wamena virus (WV), for which only the MCP gene sequence has been determined (Hyatt et al. 2002; Marsh et al. 2002), an additional MCP gene tree was constructed as described above (Table 2). The genetic divergence between the eight Thai ranaviruses and other ranaviruses was determined by aligning their MCP nucleotide sequences using the Sequence Demarcation Tool Version 1.2 (Muhire et al. 2014) with the MAFFT option implemented.
Table 2.
| Viral name | Viral abbreviation | GenBank Accession No. |
|---|---|---|
| Frog virus 3 | FV3 | AY548484 |
| Tiger frog virus | TFV | AF389451 |
| Rana grylio iridovirus | RGV | JQ654586 |
| Soft-shelled turtle iridovirus | STIV | EU627010 |
| Bohle iridovirus | BIV | KX185156 |
| German gecko ranavirus | GGRV | KP266742 |
| Ambystoma tigrinum virus | ATV | AY150217 |
| Epizootic haematopoietic necrosis virus | EHNV | FJ433873 |
| European sheatfish virus | ESV | JQ724856 |
| Common midwife toad virus | CMTV-E | JQ231222 |
| Common midwife toad virus | CMTV-NL | KP056312 |
| Testudo hermanni ranavirus | THRV-CH8/96 | KP266741 |
| Tortoise ranavirus isolate 1 | ToRV1 | KP266743 |
| Frog virus 3 isolate SSME | SSME | KJ175144 |
| Andrias davidianus ranavirus | ADRV | KC865735 |
| European catfish virus | ECV | KT989885 |
| Short-finned eel ranavirus | SERV | KX353311 |
| Ranavirus maximus | Rmax | KX574343 |
| Cod iridovirus | CoIV | KX574342 |
| Pike-perch iridovirus | PPIV | KX574341 |
| Lumpfish ranavirus isolate F140-16 | LMRV-F140-16 | MH665359 |
| Lumpfish ranavirus isolate F24-15 | LMRV-F24-15 | MH665358 |
| Lumpfish ranavirus isolate V4955 | LMRV-V4955 | MH665360 |
| Andrias davidianus ranavirus | ADRV-2010SX | KF033124 |
| Chinese giant salamander iridovirus | CGSIV-HN1104 | KF512820 |
| Common midwife toad virus | CMTV-Lv/2015 | MF004272 |
| Common midwife toad virus | CMTV-Pe/2015 | MF125269 |
| Common midwife toad virus | CMTV-Pe/2016 | MF125270 |
| Pelophylax esculentus virus | PEV | MF538627 |
| Rana catesbeiana virus isolate RC-Z | RCV-Z | MF187210 |
| Rana esculenta virus | REV | MF538628 |
| Trioceros melleri ranavirus 1 | TMRV1 | MG953519 |
| Trioceros melleri ranavirus 2 | TMRV2 | MG953520 |
| Terrapene carolina carolina ranavirus | TCCRV | MG953518 |
| Frog virus 3 | FV3-Op/2015 | MF360246 |
| Rana nigromaculata ranavirus isolate MWH421017 | RNRV-MWH421017 | MG791866 |
| Zoo ranavirus isolate 040414 | ZRV | MK227779 |
| Wamena virus | WV | MT507284 |
Results
In vitro propagation of ranaviruses
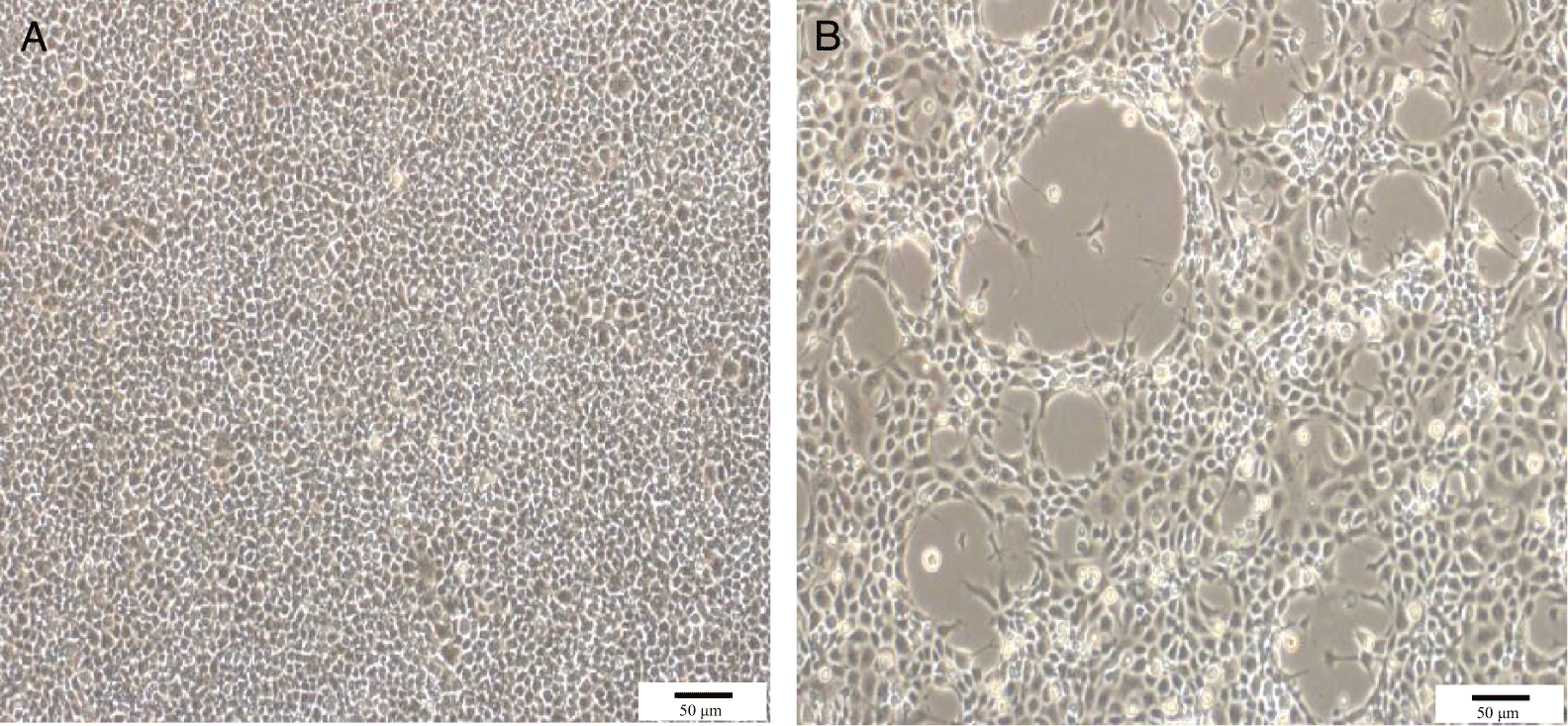
The EPC cultures inoculated with the eight Thai ranaviral isolates all displayed the same CPE characterized by enlarged, refractile cells that lysed resulting in plaques within 24 h postinoculation (Fig. 2). Complete destruction of the monolayer was observed within 48–72 h postinoculation.
Fig. 2.
TEM
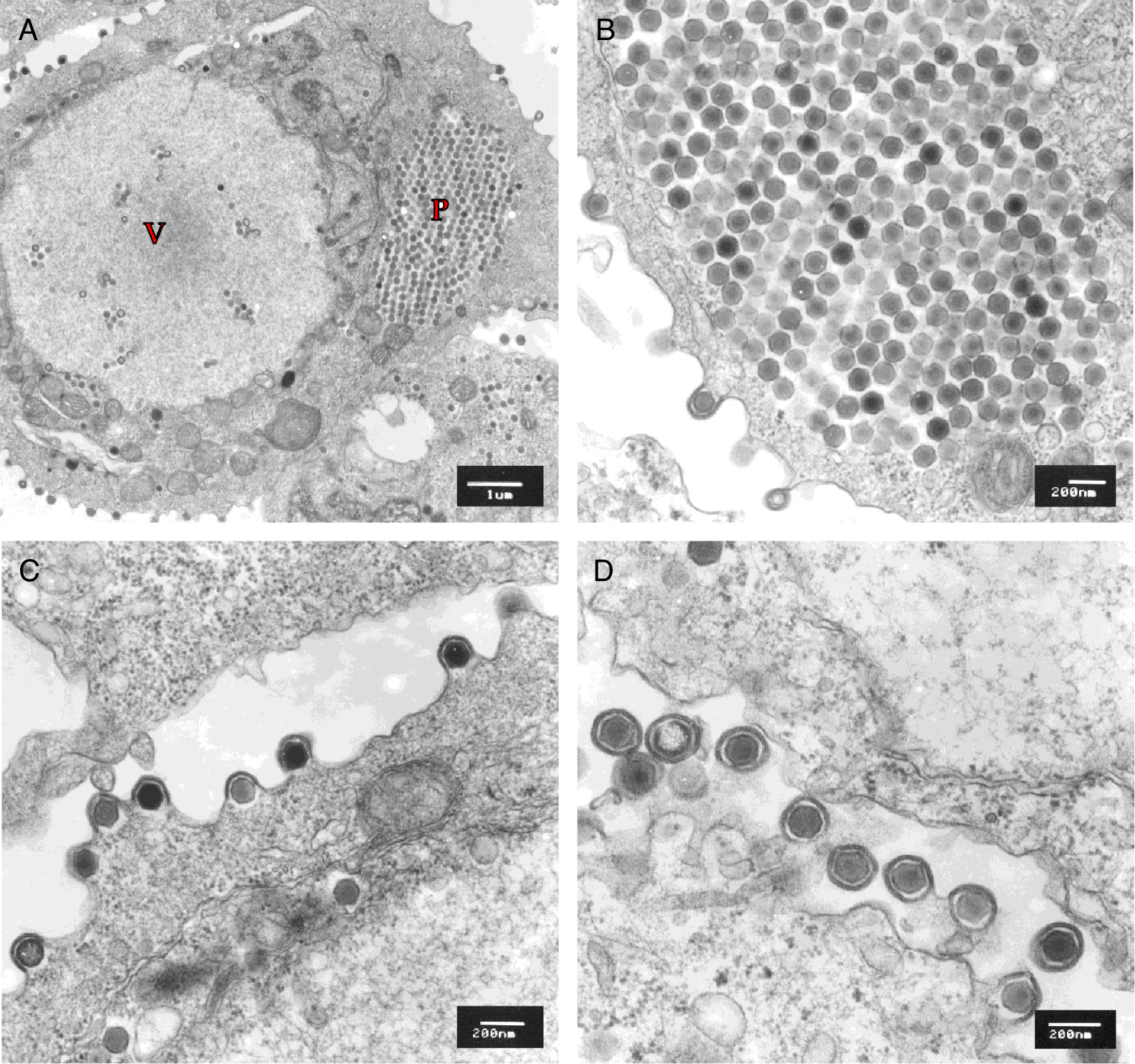
On TEM, all seven EPC cultures displayed infected cells containing abundant hexagonal nucleocapsids, each with electron-dense nucleic acid cores surrounded by a translucent zone and an outer nucleocapsid layer (Fig. 3). Mean apex–apex measurements for unenveloped and enveloped viral particles ranged from 150.2 to 158.9 nm and from 196.5 to 206.0 nm, respectively (Table 1). All isolates formed paracrystalline arrays within the cytoplasm of infected EPC cells (Figs. 3A and 3B). In all EPC cultures, viral particles were observed obtaining an envelope via budding through the plasma membrane (Figs. 3C and 3D).
Fig. 3.
Genome annotation and phylogenomic analyses
De novo assemblies of the paired-end reads generated from each of the eight sequencing libraries resulted in contiguous consensus sequences ranging in size from 105 022 to 106 226 bp with G + C contents ranging from 53.82% to 55.73% and average coverages ranging from 8341 to 17 435 reads per nucleotide (Table 3). Comparative genomic analyses predicted 100 ORFs in OMRV; 101 ORFs in AGFRV; 103 ORFS in TFV-1998, GPRV, GFRV, EABRV-11, EABRV-16, and EABRV-17; and 104 ORFS in TFV-China (Table 3). The Mauve 2.4 analysis revealed that the Chinese TFV and the eight Thai ranaviruses possessed a FV3-like ranaviral genome arrangement (Claytor et al. 2017) (Fig. S1). TFV ORFs 26 and 63, encoding hypothetical proteins, were present in the Chinese TFV and all Thai ranaviruses except OMRV (Table S2). TFV ORFs 6.5 and 59 were present in the Chinese TFV and all Thai ranaviruses except OMRV and AFGRV (Table S2). TFV ORF 54 encoding a putative nuclear calmodulin-binding protein was only present in the Chinese TFV, OMRV, and AFGRV (Table S2). TFV ORF 97, encoding a hypothetical protein, was present in the Chinese TFV and all Thai ranaviruses except AGFRV (Table S2). The Chinese TFV and Thai ranaviruses possessed a putative gene (TFV ORF 28) that was predicted to encode a protein of unknown function and was absent in all other ranaviruses (Table S2). The Chinese TFV and Thai ranaviruses were all missing FV3 ORF 30R (GenBank Accession No. NC005946) that was present in all other ranaviruses (Table S2). In addition, a single nucleotide deletion resulted in the loss of a stop codon and the merger of two ORFs (equivalent to ORFs 49L and 50L in FV3 and ORFs 51R and 52L in the Chinese TFV) in all Thai TFVs (TFV ORF 50).
Table 3.
| Viral name (abbreviation) | Isolate designation | Host | Country of origin (province) | Year of isolation | Size (kb) | No. of ORFs | % G + C | Mean coverage (reads/nt) | GenBank Accession No. |
|---|---|---|---|---|---|---|---|---|---|
| TFV-1998 | AV9803 | Tiger frog (H. tigerinus) | Thailand (Bangkok) | 1998 | 105 022 | 103 | 55.41 | 10 007 | MT512504 |
| TFV-China | TFV-China | Tiger frog (H. tigerinus) | China (Guangdong) | 2000 | 105 057 | 104 | 55 | ND | AF389451 |
Oxyeleotris marmorata ranavirus (OMRV) | D2008 | Marbled sleeper goby (Oxyeleotris marmorata) | Thailand (Nakhon Pathom) | 2000 | 105 405 | 100 | 55.73 | 10 634 | MT512502 |
Poecilia reticulata ranavirus (PPRV) | F2112 | Guppy (Poecilia reticulata) | Thailand (Samut Sakhon) | 2001 | 105 249 | 103 | 55.47 | 8888 | MT512503 |
Goldfish ranavirus (GFRV) | F0207 | Goldfish (Carassius auratus) | Thailand (Bangkok) | 2002 | 106 226 | 103 | 53.82 | 8341 | MT512501 |
Asian grass frog ranavirus (AGFRV) | D03-034 | Asian grass frog (Fejervarya limnocharis) | Thailanda (Sa Kaeo) | 2004 | 105 529 | 101 | 54.86 | 17 435 | MT512497 |
East Asian bullfrog ranavirus-2011 (EABRV-2011) | D11-067 | East Asian bullfrog (H. rugulosus) | Thailand (Phattalung) | 2011 | 105 418 | 103 | 55.03 | 9536 | MT512498 |
East Asian bullfrog ranavirus-2016 (EABRV-2016) | VD-16-006 | East Asian bullfrog (H. rugulosus) | Thailand (Ratchaburi) | 2016 | 105 206 | 103 | 55.22 | 9053 | MT512499 |
East Asian bullfrog ranavirus-2017 (EABRV-2017) | VD-17-007 | East Asian bullfrog (H. rugulosus) | Thailand (Rayong) | 2017 | 105 114 | 103 | 54.09 | 15 188 | MT512500 |
a
Frogs were recently imported from Cambodia.
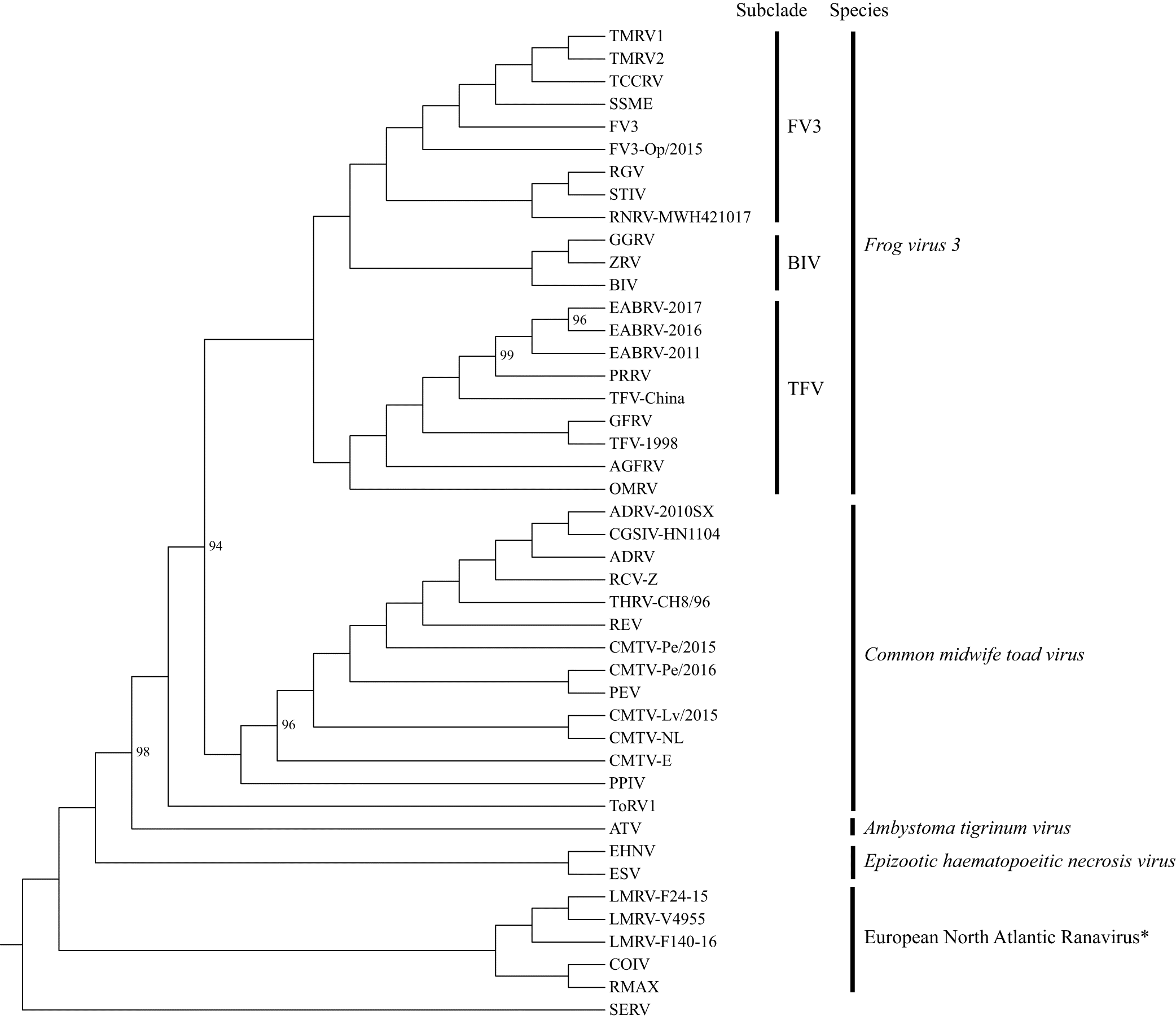
The ML analysis, based on the concatenated LCB alignment, generated a resolved and supported tree with a topology congruent to a recent analysis (Fig. 4; Stagg et al. 2020). The ML tree supported the Chinese TFV and the eight Thai ranaviruses as a unique subclade (hereafter referred to as the TFV subclade) within the larger FV3 clade. The OMRV and AGFRV isolates branched off at the base of the TFV subclade. The ML analysis, based on the MCP gene alignment, generated a similar overall topology to the tree built from the full genome (LCB) alignment; however, the resolution of this tree and its node support were significantly reduced (Fig. S3). Importantly, the MCP gene tree again supported the TFVs as a monophyletic group with the WV as the most basal branch. The MCP gene nucleotide identity of the Thai TFVs ranged from 99.8% to 100% as compared with each other and the Chinese TFV, 99.3%–99.4% when compared with the WV, and 93.7%–98.6% when compared with 35 other ranaviruses (Fig. S4).
Fig. 4.
Discussion
In this investigation, we report the complete genome sequences of eight ranaviruses isolated from cultured fish and amphibians in Thailand. The growth of the Thai ranaviruses in EPC cells facilitated their ultrastructural and phylogenomic characterization. The induction of CPE in EPC cultures (i.e., progression of coalescing round plaques) and the observed ultrastructural features of the viral particles (i.e., hexagonal viral particles within the cytoplasm and enveloped extracellular virions) were congruous with a previous report of TFV in tiger frogs in China (Weng et al. 2002) and expectations for ranaviruses (International Committee on Taxonomy of Viruses 2020). Phylogenomic analyses supported the Thai ranaviruses as a unique subclade (hereafter referred to as the TFV subclade) within the larger FV3 clade. To our knowledge, this study represents the only genetic characterization of ranaviruses in Thailand and adds to a growing body of literature on the expanding host range and potential threat ranaviruses pose to cultured fish and amphibians.
Our phylogenomic analyses strongly supported the TFV subclade as the sister group to a clade composed of three separate subclades: the Bohle iridovirus subclade (e.g., BIV, GGRV, ZRV), a second subclade with isolates from Chinese frogs (RGV, RNRV) and turtles (STIV), and a third subclade with strains from North American reptiles (TMRV1, TMRV2, TCCRV) and amphibians (FV3, SSME) and a European frog (FV3-Op). Further support for TFVs as strains of FV3 was evident by the fact that they all possess the previously defined FV3-like genome arrangement (Claytor et al. 2017). A shared derived feature (i.e., synapomorphy) that delineates TFVs from other ranaviruses, including other FV3 strains, is the acquisition of a putative gene (TFV ORF 28) that is predicted to encode a protein of unknown function. A second synapomorphy is the absence of FV3 ORF 30R (GenBank Accession No. NC005946) in TFV isolates. Our sequencing of eight Thai TFVs revealed a single nucleotide deletion resulting in the merger of two ORFs (FV3 ORFs 49L and 50L) as previously reported in the following FV3 strains: spotted salamander Maine (SSME), Rana grylio virus (RGV), and soft-shelled turtle iridovirus (STIV) (Morrison et al. 2014). In contrast, the Chinese TFV and FV3 lacked this deletion and resulting gene merger. The biological significance of these TFV gene gains, losses, and merger events remain to be determined.
The ranaviral-infected marbled sleeper goby, tadpoles, and frogs from farms in Thailand displayed cutaneous ulcers, edema (tadpoles), and no observable internal gross lesions (Kanchanakhan 1998, 2011; Kanchanakhan et al. 1999, 2004; Prasankok et al. 2005; J. Polchana, personal observation, 2017) (Fig. 1). In contrast, cutaneous ulcerative lesions were not observed in East Asian bullfrog tadpoles or frogs during TFV outbreaks on farms in China (Weng et al. 2002). The disease in cultured tadpoles in China was named “abdominal distension disease” as a result of the observed enlargement of the liver, kidney, and spleen. Petechial hemorrhages were also noted on the serosal surfaces of the enlarged internal organs. Although differences in gross lesions have been reported in cultured tadpoles and frogs on farms in China versus Thailand, the cutaneous ulceration (Thailand) and petechial hemorrhages along the serosal surfaces of enlarged internal organs (China) are all considered classic gross lesions associated with ranaviral-infected tadpoles and frogs (Miller et al. 2015). Furthermore, the tissue sections from diseased Chinese tadpoles and frogs displayed microscopic lesions consistent with ranaviral infections in amphibians including diffuse hepatocellular necrosis and focal necrosis in the kidney (Weng et al. 2002; Miller et al. 2015). Although histopathological examination of Thai TFV-infected tadpole and frog tissues has not been reported, the observation of cutaneous ulcerative lesions paired with the high prevalence of ranaviruses isolated from diseased tadpoles and frogs suggests TFV is likely a significant source of mortality in cultured amphibians in Thailand (Kanchanakhan 1998, 2011; Kanchanakhan et al. 1999, 2004; Prasankok et al. 2005). Controlled laboratory studies are needed to elucidate the role Thai TFV strains play in disease of cultured frog and fish species.
In this study, the TFV isolates from goldfish (F0207) and guppy (F2112) were recovered from overtly healthy fish during routine health surveys conducted at AAHRDD (S. Kanchanakhan, personal observation, 2001, 2002). Similarly, strains of FV3 have previously been reported from outwardly healthy fathead minnow (Pimephales promelas) and northern pike (Esox lucius) during surveillance efforts targeting viral haemorrhagic septicemia virus in the United States (Waltzek et al. 2014). Experimental challenge studies have resulted in little to no mortality in black bullhead (Ameiurus melas), bluegill (Lepomis macrochirus), channel catfish (Ictalurus punctatus), common carp (Cyprinus carpio), fathead minnow, goldfish (Carassius auratus), mosquito fish (Gambusia affinis), Nile tilapia (Oreochromis niloticus), northern pike (Esox Lucius), and pike-perch (Sander lucioperca) following exposure to FV3 isolates (Bang Jensen et al. 2009, 2011a, 2011b, Gobbo et al. 2010; Brenes et al. 2014a, 2014b). Similarly, experimental challenge studies resulted in no mortality in grass carp (Ctenopharyngodon Idella), goldfish, and mandarin fish (Siniperca chautsi) following injection of a Chinese TFV isolate (Weng et al. 2002). In contrast, farm-reared sturgeon species, including the highly endangered pallid sturgeon (Scaphirhynchus albus), appear highly susceptible to FV3 (Waltzek et al. 2014). Although few fish species appear highly susceptible to strains of FV3, at least some fish become infected and can transmit the virus to frog tadpoles under laboratory conditions (Brenes et al. 2014a).
Tiger frog virus was originally characterized from a disease episode in cultured tiger frog tadpoles and frogs in China in 2000 (He et al. 2002; Weng et al. 2002). The present study confirms that strains of TFV have been circulating in cultured fish and amphibian populations in Thailand from 1998 to 2017. Our analysis of the major capsid protein of the WV, isolated from diseased green pythons confiscated during their attempted importation into Australia from Papua New Guinea in 1998, suggests that TFVs may be circulating more widely, including in wild squamate reptiles in Oceania (Hyatt et al. 2002). Like other strains of FV3, TFVs are promiscuous pathogens capable of infecting three classes of vertebrates including bony fish, amphibians, and reptiles (Chinchar and Waltzek 2014). Experimental studies have demonstrated interclass transmission can occur by cohabitation of FV3-infected Cope’s treefrog tadpoles (Hyla chrysoscelis) with naïve red-eared sliders (Trachemys scripta elegans) and cohabitation of FV3-infected mosquitofish (Gambusia affinis) or FV3-infected red-eared sliders with naïve Cope’s treefrog tadpoles (Brenes et al. 2014a). In Thailand, opportunities for interclass transmission of TFVs may occur at large pet markets where tadpoles and frogs are housed in open containers next to ornamental fishes. The aquaculture practice of feeding juvenile frogs to increase the growth of large predatory ornamental fishes may have also contributed to the spread of TFVs in Thailand (S. Kanchanakhan, personal observation, 2016).
Continued TFV surveillance efforts are needed to monitor any impact these ranaviruses may have on Thai aquatic animal industries. Improved biosecurity programs should be implemented (e.g., discontinuing the practice of feeding live juvenile frogs to predatory fishes) to reduce the impact of infectious diseases on Thai farms and pet markets. Although TFVs have only been detected in Asia and likely in Oceania, their promiscuous nature coupled with the largely unregulated international trade in live animals raises concerns that they might emerge in more distant ranaculture and aquaculture markets or wildlife populations (Picco and Collins 2008; Schloegel et al. 2009; Kolby et al. 2014).
Acknowledgements
We would like to thank Yuwanda Yeeka at the Aquatic Animal Research and Development Division, Department of Fisheries, Thailand, for her assistance in cell culture.
References
Bang Jensen B, Ersbøll AK, and Ariel E. 2009. Susceptibility of pike Esox lucius to a panel of Ranavirus isolates. Diseases of Aquatic Organisms, 83(3): 169–179.
Bang Jensen B, Holopainen R, Tapiovaara H, and Ariel E. 2011a. Susceptibility of pike-perch Sander lucioperca to a panel of ranavirus isolates. Aquaculture, 313(1–4): 24–30.
Bang Jensen B, Reschova S, Cinkova K, Ariel E, and Vesely T. 2011b. Common carp (Cyprinus carpio) and goldfish (Carassius auratus) were not susceptible to challenge with ranavirus under certain challenge conditions. Bulletin of the European Association of Fish Pathologists, 31(3): 112–118.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology, 19(5): 455–477.
Bennett M, Tu SL, Upton C, McArtor C, Gillett A, Laird T, et al. 2017. Complete genomic characterisation of two novel poxviruses (WKPV and EKPV) from western and eastern grey kangaroos. Virus Research, 242: 106–121.
Besemer J, Lomsadze A, and Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Research, 29(12): 2607–2618.
Brenes R, Gray MJ, Waltzek TB, Wilkes RP, and Miller DL. 2014a. Transmission of ranavirus between ectothermic vertebrate hosts. PLoS ONE, 9(3): e92476.
Brenes R, Miller DL, Waltzek TB, Wilkes RP, Tucker JL, Chaney JC, et al. 2014b. Susceptibility of fish and turtles to three ranaviruses isolated from different ectothermic vertebrate classes. Journal of Aquatic Animal Health, 26(2): 118–126.
Chen ZX, Zheng JC, and Jiang YL. 1999. A new iridovirus isolated from soft-shelled turtle. Virus Research, 63: 147–151.
Chinchar VG, and Waltzek TB. 2014. Ranaviruses: not just for frogs. PLoS Pathogens, 10(1): e1003850.
Chinchar VG, Hick P, Ince IA, Jancovich JK, Marschang R, Qin Q, et al. 2017. ICTV virus taxonomy profile: Iridoviridae. Journal of General Virology, 98: 890–891.
Claytor SC, Subramaniam K, Landrau-Giovannetti N, Chinchar VG, Gray MJ, Miller DL, et al. 2017. Ranavirus phylogenomics: signatures of recombination and inversions among bullfrog ranaculture isolates. Virology, 511: 330–343.
Darling AC, Mau B, Blattner FR, and Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Research, 14(7): 1394–1403.
Department of Fisheries Thailand. 2019a. Fisheries statistics of Thailand 2017. Fishery Statistics Analysis and Research Group, Fisheries Development Policy and Strategy Division, Department of Fisheries, Bangkok, Thailand. Vol. 9, pp. 13–14.
Department of Fisheries Thailand. 2019b. Statistics of freshwater aquaculture production 2017. Fishery Statistics Analysis and Research Group, Fisheries Development Policy and Strategy Division, Department of Fisheries, Bangkok, Thailand. Vol. 4, 25 p.
Duffus ALJ, and Cunningham AA. 2010. Major disease threats to European amphibians. Herpetological Journal, 20: 117–127.
Duffus ALJ, Waltzek TB, Stöhr AC, Allender MC, Gotesman M, Whittington R, et al. 2015. Distribution and host range of ranaviruses. In Ranaviruses. Edited by MJ Gray and VG Chinchar. Springer, Cham, Switzerland. pp. 9–58.
Earl JE, and Gray MJ. 2014. Introduction of ranavirus to isolated wood frog populations could cause local extinction. EcoHealth, 11: 581–592.
Food and Agriculture Organization of the United Nations. 2020. Fishery and aquaculture country profiles, Thailand, country profile fact sheets. FAO Fisheries and Aquaculture Department, Rome, Italy [online]: Available from fao.org/fishery/facp/THA/en.
Gobbo F, Cappellozza E, Pastore MR, and Bovo G. 2010. Susceptibility of black bullhead Ameiurus melas to a panel of ranavirus isolates. Diseases of Aquatic Organisms, 90(3): 167–174.
He JG, Lü L, Deng M, He HH, Weng SP, Wang XH, et al. 2002. Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology, 292(2): 185–197.
Hyatt AD, Williamson M, Coupar BE, Middleton D, Hengstberger SG, Gould AR, et al. 2002. First identification of a ranavirus from green pythons (Chondropython viridis). Journal of Wildlife Diseases, 38(2): 239–252.
International Committee on Taxonomy of Viruses. 2020. ICTV virus taxonomy profile: Iridoviridae, Genus Ranavirus [online]: Available from talk.ictvonline.org/ictv-reports/ictv_online_report/dsdna-viruses/w/iridoviridae/616/genus-ranavirus.
Kanchanakhan S. 1998. An ulcerative disease of the cultured tiger frog, Rana tigrina, in Thailand: virological examination. Aquatic Animal Health Research Institute (AAHRI) Newsletter, 7: 1–2.
Kanchanakhan S. 2011. Ranaviruses in frogs and fish in Southeast Asia. In Joint Meeting of Ichthyologists and Herpetologists (JMIH), Minneapolis, Minnesota, 6–11 July 2011.
Kanchanakhan S, Saduakdee U, and Chinabut S. 1999. Isolation and characterization of a new virus from ulcerative diseased tiger frog, Rana tigrina Cantor. In 37th Kasetsart University Annual Conference, Bangkok, Thailand, 3–5 February 1999. pp. 133–138.
Kanchanakhan S, Polchana J, Nuanchan T, and Tantavanijt S. 2004. Isolation of characterisation of ranavirus from diseased frog, Rana spp., imported from Cambodia. Inland Aquatic Animal Health Research Institute, Bangkok, Thailand, abstract.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12): 1647–1649.
Koda SA, Subramaniam K, Francis-Floyd R, Yanong RP, Frasca S Jr, Groff JM, et al. 2018. Phylogenomic characterization of two novel members of the genus Megalocytivirus from archived ornamental fish samples. Diseases of Aquatic Organisms, 130(1): 11–24.
Kolby JE, Smith KM, Berger L, Karesh WB, Preston A, Pessier AP, et al. 2014. First evidence of amphibian chytrid fungus (Batrachochytrium dendrobatidis) and ranavirus in Hong Kong amphibian trade. PLoS ONE, 9(3): e90750.
Langmead B, and Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nature Methods, 9(4): 357–359.
Marsh IB, Whittington RJ, O’Rourke B, Hyatt AD, and Chisholm O. 2002. Rapid differentiation of Australian, European and American ranaviruses based on variation in major capsid protein gene sequence. Molecular and Cellular Probes, 16(2): 137–151.
Mazzoni R, de Mesquita AJ, Fleury LF, de Brito WM, Nunes IA, Robert J, et al. 2009. Mass mortality associated with a frog virus 3-like Ranavirus infection in farmed tadpoles Rana catesbeiana from Brazil. Diseases of Aquatic Organisms, 86(3): 181–191.
Miller DL, Rajeev S, Gray MJ, and Baldwin CA. 2007. Frog virus 3 infection, cultured American bullfrogs. Emerging Infectious Diseases, 13(2): 342–343.
Miller DL, Gray MJ, and Storfer AT. 2011. Ecopathology of ranaviruses infecting amphibians. Viruses, 3(11): 2351–2373.
Miller DL, Pessier AP, Hick P, and Whittington RJ. 2015. Comparative pathology of ranaviruses and diagnostic techniques. In Ranaviruses. Edited by MJ Gray and VG Chinchar. Springer, Cham, Switzerland. pp. 171–208.
Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, et al. 2013. Using tablet for visual exploration of second-generation sequencing data. Briefings in Bioinformatics, 14(2): 193–202.
Morrison EA, Garner S, Echaubard P, Lesbarrères D, Kyle CJ, and Brunetti CR. 2014. Complete genome analysis of a frog virus 3 (FV3) isolate and sequence comparison with isolates of differing levels of virulence. Virology Journal, 11: 46.
Muhire BM, Varsani A, and Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE, 9(9): e108277.
Nguyen LT, Schmidt HA, von Haeseler A, and Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32(1): 268–274.
Peace A, O’Regan SM, Spatz JA, Reilly PN, Hill RD, Carter ED, et al. 2019. A highly invasive chimeric ranavirus can decimate tadpole populations rapidly through multiple transmission pathways. Ecological Modelling, 410: 108777.
Picco AM, and Collins JP. 2008. Amphibian commerce as a likely source of pathogen pollution. Conservation Biology, 22(6): 1582–1589.
Prasankok P, Chutmongkonkul M, and Kanchanakhan S. 2005. Characterisation of iridovirus isolated from diseased marbled sleepy goby, Oxyeleotris marmoratus. In Diseases in Asian aquaculture V. Edited by P Walker, R Lester, and MG Bondad-Reantaso. Fish Health Section, Asian Fisheries Society, Manila, Philippines. pp. 197–206.
Price SJ, Garner TWJ, Nichols RA, Balloux F, Ayres C, de Alba AMC, et al. 2014. Collapse of amphibian communities due to an introduced Ranavirus. Current Biology, 24: 2586–2591.
Schloegel LM, Picco AM, Kilpatrick AM, Daviesd AJ, Hyatte AD, and Daszak P. 2009. Magnitude of the US trade in amphibians and presence of Batrachochytrium dendrobatidis and ranavirus infection in imported North American bullfrogs (Rana catesbeiana). Biological Conservation, 142(7): 1420–1426.
Stagg HEB, Guðmundsdóttir S, Vendramin N, Ruane NM, Sigurðardóttir H, Christiansen DH, et al. 2020. Characterization of ranaviruses isolated from lumpfish Cyclopterus lumpus L. in the North Atlantic area: proposal for a new ranavirus species (European North Atlantic Ranavirus). Journal of General Virology, 101(2): 198–207.
Tan WG, Barkman TJ, Gregory CV, and Essani K. 2004. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae). Virology, 323(1): 70–84.
Tcherepanov V, Ehlers A, and Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics, 7: 150.
Teacher AGF, Cunningham AA, and Garner TWJ. 2010. Assessing the long‐term impact of ranavirus infection in wild common frog populations. Animal Conservation, 13: 514–522.
Waltzek TB, Miller DL, Gray MJ, Drecktrah B, Briggler JT, MacConnell B, et al. 2014. New disease records for hatchery-reared sturgeon. I. Expansion of Frog virus 3 host range into Scaphirhynchus albus. Diseases of Aquatic Organisms, 111(3): 219–227.
Weng SP, He JG, Wang XH, Lü L, Deng M, and Chan SM. 2002. Outbreaks of an iridovirus disease in cultured tiger frog, Rana tigrina rugulosa, in southern China. Journal of Fish Diseases, 25: 423–427.
World Organization for Animal Health. 2019. Chapter 2.3.1. Infection with epizootic haematopoietic necrosis virus. In Manual of diagnostic tests for aquatic animals. OIE, Paris, France [online]: Available from oie.int/index.php?id=2439&L=0&htmfile=chapitre_ehn.htm.
Zhang QY, Xiao F, Li ZQ, Gui JF, Mao J, and Chinchar VG. 2001. Characterization of an iridovirus from the cultured pig frog, Rana grylio, with lethal syndrome. Diseases of Aquatic Organisms, 48(1): 27–36.
Supplementary materials
Supplementary Material 1 (PDF / 743 KB)
- Download
- 743.21 KB
Supplementary Material 2 (DOCX / 60.2 KB)
- Download
- 60.28 KB
Supplementary Material 3 (PDF / 468 KB)
- Download
- 468.59 KB
Supplementary Material 4 (PDF / 331 KB)
- Download
- 331.01 KB
Information & Authors
Information
Published In

FACETS
Volume 5 • Number 1 • January 2020
Pages: 963 - 979
Editor: Ellen Ariel
History
Received: 4 June 2020
Accepted: 9 September 2020
Version of record online: 3 December 2020
Notes
This paper is part of a Collection titled “Ranavirus research: 10 years of global collaboration.”
Copyright
© 2020 Sriwanayos et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper and in the Supplementary Material.
Key Words
Sections
Subjects
Authors
Author Contributions
PS, KS, VLP, and TBW conceived and designed the study.
PS, KS, NKS, and KI performed the experiments/collected the data.
PS, KS, VLP, and TBW analyzed and interpreted the data.
SK, JP, and TBW contributed resources.
All drafted or revised the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Preeyanan Sriwanayos, Kuttichantran Subramaniam, Natalie K. Stilwell, Kamonchai Imnoi, Vsevolod L. Popov, Somkiat Kanchanakhan, Jaree Polchana, and Thomas B. Waltzek. 2020. Phylogenomic characterization of ranaviruses isolated from cultured fish and amphibians in Thailand. FACETS.
5(1): 963-979. https://doi.org/10.1139/facets-2020-0043
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Phylogenomic Characterization of Ranavirus Isolated from Wild Smallmouth Bass (Micropterus dolomieu)
2. Emerging threat of ranavirus: prevalence, genetic diversity, and climatic drivers of Ranavirus (Iridoviridae) in ectothermic vertebrates of Asia
3. Emerging importance of bighead catfish (Clarias macrocephalus) and north African catfish (C. gariepinus) as a bioresource and their genomic perspective
4. Recent insights into aquatic viruses: Emerging and reemerging pathogens, molecular features, biological effects, and novel investigative approaches
5. Molecular Confirmation of Ranavirus Infection in Amphibians From Chad, Africa
