Global patterns of ranavirus detections
Abstract
Ranaviruses are emerging pathogens of poikilothermic vertebrates. In 2015 the Global Ranavirus Reporting System (GRRS) was established as a centralized, open access, online database for reports of the presence (and absence) of ranavirus around the globe. The GRRS has multiple data layers (e.g., location, date, host(s) species, and methods of detection) of use to those studying the epidemiology, ecology, and evolution of this group of viruses. Here we summarize the temporal, spatial, diagnostic, and host-taxonomic patterns of ranavirus reports in the GRRS. The number, distribution, and host diversity of ranavirus reports have increased dramatically since the mid 1990s, presumably in response to increased interest in ranaviruses and the conservation of their hosts, and also the availability of molecular diagnostics. Yet there are clear geographic and taxonomic biases among the reports. We encourage ranavirus researchers to add their studies to the portal because such collation can provide collaborative opportunities and unique insights to our developing knowledge of this pathogen and the emerging infectious disease that it causes.
The Global Ranavirus Reporting System
The Global Ranavirus Reporting System (GRRS; brunnerlab.shinyapps.io/GRRS_Interactive/) was designed as a central, open-access, online database in which to report and track the occurrence of ranavirus presence or absence around the world (Duffus and Olson 2011). Globally compiled detections and nondetections of ranaviruses can provide novel insights into pathogen emergence, host range, and susceptibility. In addition to occurrences, the GRRS is an archive of ranavirus field and diagnostic studies that include additional project aims and details such as detection methods.
Since its initiation in 2015 we have been working to populate the database with records of ranavirus detections. We have focused our efforts on published accounts of ranavirus occurrence, although the GRRS also includes dissertations, seminars, and unpublished accounts, and more are welcome. As of this writing there are 206 studies or publications in the GRRS that provide data on the occurrence of ranaviruses in one or more taxa (Fig. 1). This is, of course, a fraction of publications on ranaviruses more broadly (e.g., phylogenetics, experimental studies; see Blaustein et al. (2018) for a review of experimental studies) and, we expect, is far from inclusive of all location or occurrence data hiding in field records and on laboratory computers. One goal of this note is to encourage the broader community to add their unpublished reports of ranavirus to the GRRS. Another is to ask researchers to find their references and offer corrections or refinements to the information in the database. We therefore begin by describing the data in the GRRS and noting some common issues that are likely to be important for those using the GRRS for various analyses.
Fig. 1.
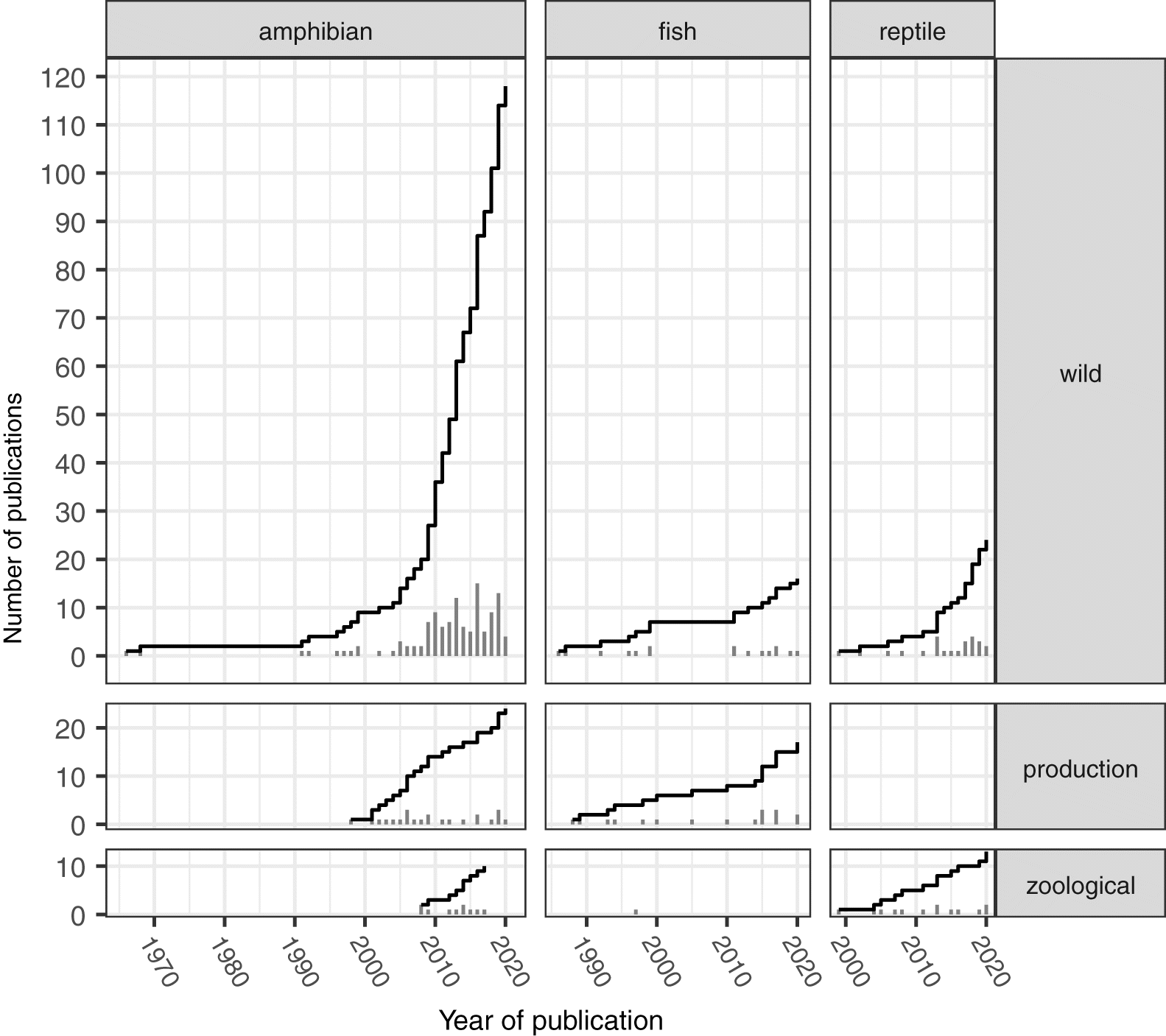
The GRRS represents a unique tool for researchers and policy makers. Whereas prior publications have described, for instance, the taxonomic breadth of ranavirus hosts (Duffus et al. 2015), the data in the GRRS system provide additional context and facilitate additional analyses. In particular, the GRRS offers a broad geographic and temporal perspective on the occurrence of ranaviruses and the settings in which ranaviruses have been found. We therefore highlight several interesting patterns in the reports in the GRRS and suggest possible interpretations and hypotheses. We hope that these will inspire further research and more reporting with greater precision.
Data in the GRRS
The GRRS includes information on the location and date of sample collection; species, life stages, and type of population sampled (i.e., wild, production, or zoological); and information on the numbers of samples screened for infection, confirmed infected, confirmed diseased if relevant, and the type of diagnostic methods employed. Samples include both animal tissues (e.g., liver or tail clip) as well as swabs. They do not yet include environmental DNA or other samples not clearly linked to an individual host. When DNA sequencing data had been submitted to GenBank, the accession number is also included. Each record includes the publication’s title, journal or reporting outlet, volume, and pages (or contact information for unpublished records) so that a user may trace records back to the original publication or source. See the GRRS site for additional metadata.
The data in the GRRS are presented as an interactive map, where clicking on a record (one row in the database is one dot on the map) provides more information about that record. The map can be zoomed and moved. The records are subsettable by population type and taxa. All data can be freely downloaded. The database is organized so that each row represents a unique combination of location, date, or species from a study, where these information were available. Thus a single publication may be represented in the database by one or a series of rows (and points on the map). Again, the database includes both detections and nondetections; both show up on the map. All but seven publications, however, include at least one detection and just 41% of all records are nondetections.
While the GRRS represents a uniquely comprehensive database on the host and geographic distribution of ranaviruses, it is important to note a few important limitations to the underlying records and how we dealt with them. These limitations stem, in large measure, from the fact that many publications in the GRRS focus on the biology of ranaviruses and their hosts (e.g., virology, phylogenetics, conservation) rather than on populating a database of occurrences. We hope, however, that readers will consider that their publications may be used in unexpected ways and thus provide greater detail about their collections and results in the future. We also encourage readers to contact us to improve or correct our best guesses for those publications already in the GRRS.
One common issue was simply vague descriptions of the locations or timing of sample collections. For instance, the virus that became the type species of Ranavirus, Frog virus 3 (FV3), was described as coming from “Wisconsin-Minnesota frogs” (Rana pipiens) in Granoff et al. (1965) and as “derived from an inclusion-containing Wisconsin Lucke tumor” in Rafferty (1965), presumably from a biological supplier collecting from the wild, and neither provided a date of collection. In cases with vague location information we used the midpoint or centroid (e.g., of the state of Wisconsin) or the nearest mentioned landmark (e.g., a city) as the location. When dates were not provided we have used 1 January of the year of publication and when only a month or season was provided we have done our best to find a reasonable midpoint. Note that missing location and temporal data were especially common in earlier records and, perhaps intentionally, also in records from production facilities. Other reports, especially those with a more ecological focus, often provide great detail on locations, dates, and numbers. Hence the degree of accuracy and precision in location data in the GRRS is quite variable.
Descriptions of locations for imported animals offer unique challenges; should we use the source’s or importer’s location? In some cases the source was much less clear than the destination (e.g., reptiles imported into Germany from “Asia”) and so we would use the more precise location. In other cases (e.g., the case of newts imported into Germany from a specific location in Iraq; Stöhr et al. 2013a) we were able to use the location of the source, at least when the event or detection was recent (within weeks to a few months) of importation. When animals were maintained in a facility for longer periods we would consider this facility the location of record (e.g., as part of a collection or for research; Stöhr et al. 2013b).
Second, some care is also required in interpreting taxonomic designations of hosts as nomenclature has changed and, especially for some amphibians, remains contentious. We have left species epithets as written or provided in the publication, but also provided a “translation” to a common set of names of genera so that a single species is not counted multiple times (e.g., Lithobates –> Rana).
It is also important to note that taxonomy of the genus Ranavirus, and indeed the family Iridoviridae, has advanced a great deal in the last decade. For instance, the GRRS originally listed five species—FV3, Ambystoma tigrinum virus, Bohle iridovirus virus (BIV), European catfish virus (ECV), and Santee-Couper virus as well as “other”—which was current as of 2015, but the latest taxonomic revision removes BIV and ECV and adds Singapore grouper iridovirus and Common midwife toad virus (ICTV 2020). Some manuscripts also propose new ranavirus species (e.g., European North Atlantic Ranavirus; Stagg et al. 2020). Moreover, the evidence used for species designations has changed over the years (e.g., from restriction fragment length profiles to sequences of portions of key genes to whole genome sequencing). We therefore simply list species as identified in the publication and encourage users interested in ranavirus species designations to review the original papers.
We have also attempted to record the type of population from which samples were collected, although this sometimes involves a degree of interpretation. For instance we have designated the original records of FV3 as having come from the wild, even though the animals were obtained from a biological supplier, as we know suppliers tended to collect from the wild and this designation seemed more appropriate than a production setting (e.g., a ranaculture facility). For some taxa the distinction between wild and production settings is in the eye of the beholder (e.g., the Chinese giant salamander (Andrias davidianus) or American bullfrogs (Lithobates catesbeianus) that are “ranched” in constructed or semi-natural ponds intended for production, but essentially unmanaged and open to immigration or emigration). In general, “production” was interpreted as being raised or held with the intent of distribution. “Zoological” populations include zoos, private collections, and research populations or colonies not intended for distribution.
Users should be cautious interpreting data on the number of individuals involved or tested. Often this information was simply not provided, at least in a way that can be easily parsed. Perhaps more importantly, different types of research involve very different intensities of sampling (e.g., virus identification from one or a few carcasses vs. a broad epidemiological survey with hundreds of samples). We therefore avoid making comparisons in this manuscript about the impact of ranavirus or number of animals involved in different settings or taxa.
Lastly, it is important to note that the GRRS is not associated with the OIE (World Organization for Animal Health), which lists ranavirus as notifiable diseases (Schloegel et al. 2010). Uploading records to the GRRS does not satisfy OIE and national reporting practices for notifiable infections. Please refer to the reporting rules of the country in which you work and see the OIE’s World Animal Health Information System (OIE 2020).
Patterns in reporting
Since the original publications by Granoff et al. (1965) and Rafferty (1965) describing FV3 isolated from wild amphibians, ranavirus reports were rare for decades. Only in the late 1990s did the number of reports begin to grow substantially (Fig. 1). Overall, there are far more reports from amphibians than fish and reptiles and from the wild than production or zoological settings. To a large degree this probably reflects an increased interest in amphibian diseases as threats contributing to global declines in amphibian populations (Daszak et al. 1999). Indeed, the increase in reports from amphibians in the GRRS mirrors an overall increase in publications about amphibian pathogens (Blaustein et al. 2018). Moreover, most reports from the mid 1980s through the early 2000s focused on mortality events, although there has been a clear trend towards routine screening in wild populations in recent years (Fig. 2). To the extent that ranaviruses were found in fish and reptiles in the wild, it tended to be in sport fish in recreation sites and turtles in reserves.
Fig. 2.
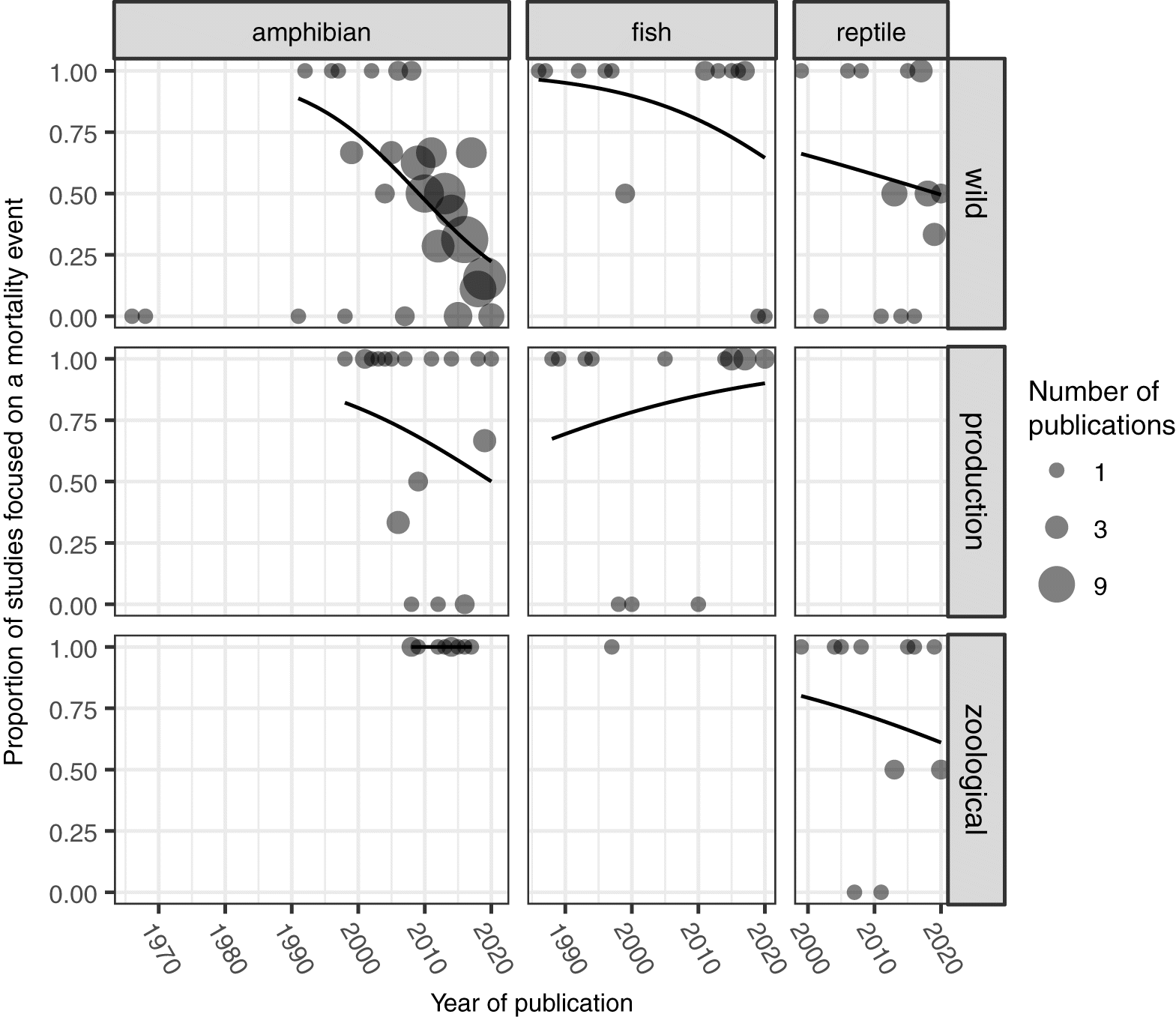
Reports in production and zoological settings tended to stem from mortality events rather than routine screening (Fig. 2). Reports from amphibians in production settings included bullfrogs, Rana catesbeiana (n = 7 publications), but also single records from other ranids and two salamanders. Interestingly, there are no current GRRS reports of reptiles in production settings. Reports from fish in production settings were limited to single instances per species. Surprisingly there was only one instance of a ranavirus from fish in a zoological setting. The reason for this apparent absence is unclear given the enormous numbers of ornamental fish kept and traded around the globe (Whittington and Chong 2007; Smith et al. 2017).
Patterns in host taxonomy
We also see that most diversity at the genus level in the GRRS is found in amphibians, primarily anurans (n = 63), but fish (n = 27) and reptile (n = 34) genera are also well represented (Table 1). This representation, however, is a fraction of the diversity within these classes. For instance, the 63 amphibian genera in the GRRS represents just 9.8% of the 646 currently recognized (AmphibiaWeb 2020). Moreover, we are aware of no reports of ranaviruses in caecilians. Presumably this reflects limited search efforts more than a restricted host range. Similar to the numbers of reports, most host diversity in the GRRS comes from wild populations rather than production or zoological settings. This is perhaps not surprising in production settings, which involves relatively few host taxa overall, but zoos and private collections comprise a great deal of diversity and so the relative absence of reports in these settings suggests under-reporting. Moreover, this collective under-reporting should cause us pause before we use the diversity of hosts from which ranaviruses have been reported as a measure of host range more generally.
Table 1.
| Class | Order | Family | Wild | Production | Zoological | Total |
|---|---|---|---|---|---|---|
| Amphibian | Anura | Alytidae | 1 | 0 | 1 | 2 |
| Batrachylidae | 1 | 0 | 0 | 1 | ||
| Bombinatoridae | 1 | 0 | 0 | 1 | ||
| Bufonidae | 5 | 0 | 2 | 7 | ||
| Calyptocephalellidae | 2 | 1 | 0 | 3 | ||
| Centrolenidae | 3 | 0 | 0 | 3 | ||
| Craugastoridae | 2 | 0 | 0 | 2 | ||
| Dendrobatidae | 1 | 0 | 3 | 4 | ||
| Dicroglossidae | 0 | 1 | 0 | 1 | ||
| Eleutherodactylidae | 2 | 0 | 0 | 2 | ||
| Hylidae | 7 | 0 | 1 | 8 | ||
| Leptodactylidae | 2 | 0 | 0 | 2 | ||
| Limnodynastidae | 1 | 0 | 0 | 1 | ||
| Microhylidae | 2 | 0 | 0 | 2 | ||
| Pelobatidae | 1 | 0 | 0 | 1 | ||
| Pelodryadidae | 1 | 0 | 1 | 2 | ||
| Phyllomedusidae | 1 | 0 | 0 | 1 | ||
| Pipidae | 1 | 1 | 0 | 2 | ||
| Pyxicephalidae | 0 | 0 | 1 | 1 | ||
| Ranidae | 2 | 1 | 2 | 5 | ||
| Rhacophoridae | 0 | 0 | 1 | 1 | ||
| Scaphiopodidae | 2 | 0 | 0 | 2 | ||
| Telmatobiidae | 1 | 0 | 0 | 1 | ||
| Urodela | Ambystomatidae | 1 | 1 | 0 | 2 | |
| Cryptobranchidae | 2 | 1 | 1 | 4 | ||
| Plethodontidae | 7 | 0 | 0 | 7 | ||
| Salamandridae | 8 | 0 | 0 | 8 | ||
| Fish | Acipenseriformes | Acipenseridae | 0 | 1 | 0 | 1 |
| Actinopterygii | Anguillidae | 0 | 1 | 0 | 1 | |
| Gadidae | 1 | 0 | 0 | 1 | ||
| Nemacheilidae | 0 | 1 | 0 | 1 | ||
| Pomacentridae | 0 | 1 | 0 | 1 | ||
| Scophthalmidae | 0 | 1 | 0 | 1 | ||
| Centrarchiformes | Oplegnathidae | 1 | 0 | 0 | 1 | |
| Percichthyidae | 0 | 1 | 0 | 1 | ||
| Terapontidae | 0 | 1 | 0 | 1 | ||
| Cichliformes | Cichlidae | 1 | 0 | 1 | 2 | |
| Cypriniformes | Cobitidae | 1 | 0 | 0 | 1 | |
| Cyprinidae | 1 | 1 | 0 | 2 | ||
| Gasterosteiformes | Gasterosteidae | 1 | 0 | 0 | 1 | |
| Gobiiformes | Butidae | 1 | 1 | 0 | 2 | |
| Perciformes | Centrarchidae | 1 | 0 | 0 | 1 | |
| Latidae | 1 | 0 | 0 | 1 | ||
| Lutjanidae | 1 | 0 | 0 | 1 | ||
| Percidae | 1 | 1 | 0 | 2 | ||
| Serranidae | 1 | 1 | 0 | 2 | ||
| Sparidae | 1 | 0 | 0 | 1 | ||
| Salmoniformes | Salmonidae | 0 | 1 | 0 | 1 | |
| Scorpaeniformes | Cyclopteridae | 1 | 1 | 0 | 2 | |
| Siluriformes | Ictaluridae | 2 | 0 | 0 | 2 | |
| Siluridae | 1 | 1 | 0 | 2 | ||
| Reptile | Squamata | Agamidae | 4 | 0 | 3 | 7 |
| Anguidae | 1 | 0 | 1 | 2 | ||
| Chamaeleonidae | 0 | 0 | 1 | 1 | ||
| Colubridae | 1 | 0 | 0 | 1 | ||
| Dactyloidae | 0 | 0 | 1 | 1 | ||
| Gekkonidae | 0 | 0 | 1 | 1 | ||
| Iguanidae | 0 | 0 | 1 | 1 | ||
| Lacertidae | 1 | 0 | 0 | 1 | ||
| Phrynosomatidae | 1 | 0 | 0 | 1 | ||
| Pythonidae | 1 | 0 | 1 | 2 | ||
| Scincidae | 0 | 0 | 1 | 1 | ||
| Testudines | Chelydridae | 1 | 0 | 0 | 1 | |
| Emydidae | 8 | 0 | 1 | 9 | ||
| Kinosternidae | 2 | 0 | 0 | 2 | ||
| Testudinidae | 1 | 0 | 3 | 4 | ||
| Trionychidae | 2 | 0 | 0 | 2 |
Ranavirus geography
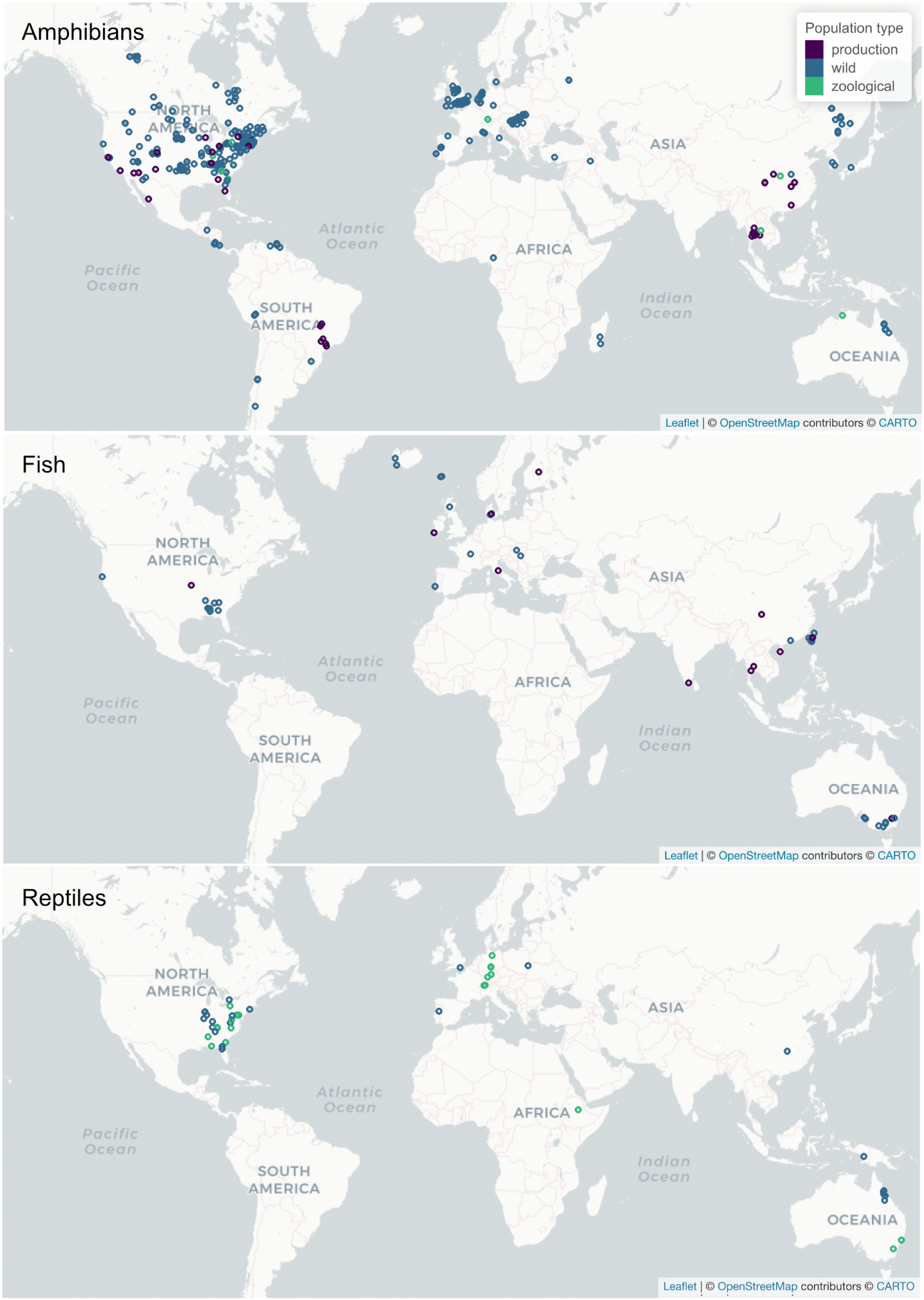
The maps of ranavirus reports (Fig. 3) illustrate a common pattern in disease detection: one finds infections where one looks. There is a clear bias towards reports in North America, Europe, and Southeast Asia. Reports from South America are sporadic and there are only three from Africa, including Madagascar. It is also worth noting that most reports of ranavirus are not from diversity hotspots (e.g., there are relatively few reports from amphibians in the tropics). Hence, one would expect that we have yet to discern the true geographic distribution (or host range) of ranaviruses. Most reports from production settings (e.g., ranaculture, aquaculture) are from Southeast Asia (i.e., China with 11, Thailand with four), but also from the United States (with nine reports) and Brazil (with an additional four).
Fig. 3.
Dominant modes of detection
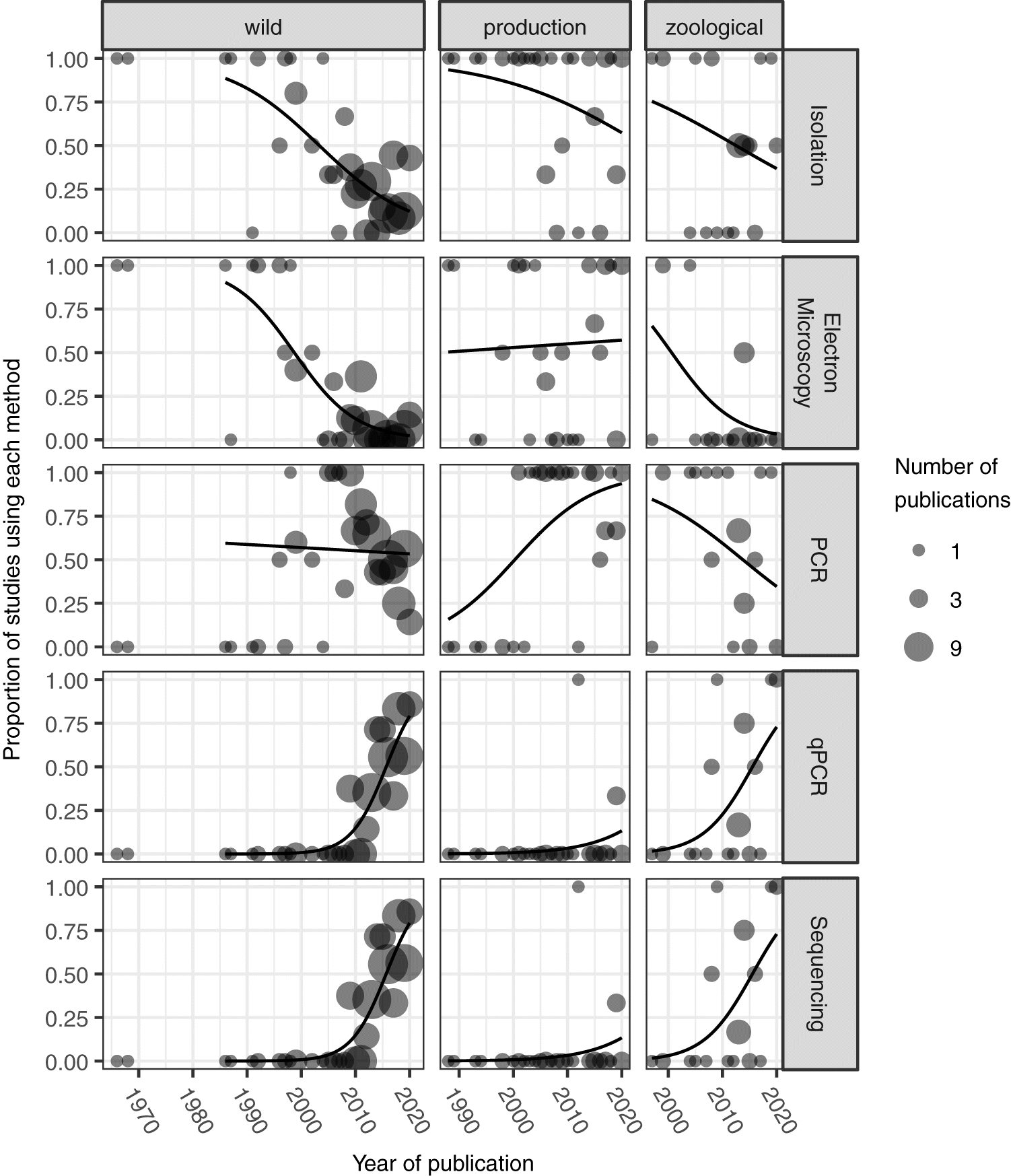
As the available diagnostic methods for detecting ranavirus have grown, their representation in the literature has changed (Fig. 4). Virus isolation in culture has continued through at least the early 2010s, but has increasingly been replaced by molecular methods. Electron microscopy is increasingly rare except in production settings. Similarly, traditional polymerase chain reaction (PCR) has largely been supplanted by quantitative, primarily Taqman, real-time PCR (qPCR). We suspect the broader adoption and relative ease of screening samples with (q)PCR methods is partially responsible for the increased routine testing of populations for ranaviruses and other pathogens (Fig. 2). Gene sequencing has also become increasingly common in certain contexts, presumably as prices have decreased, access increased, and researchers have realized the biological significance of ranaviral species and genotypes. Much of this sequencing focuses on the major capsid protein gene and a few other genes, which may be useful in differentiating among ranavirus species (Tidona et al. 1998). However, with increasing evidence of inversions and recombination among strains (e.g., Claytor et al. 2017) we hope to see more and deeper sequencing to better characterize the ranaviruses being detected. We also hope that more researchers attempt to isolate ranaviruses. Virus isolates are the raw materials for subsequent experiments and viruses are known to evolve and attenuate with repeated passage in culture (Ebert 1998). All that said, reports of ranavirus occurrence are interesting and useful even without these additional data. We encourage researchers to add their data to the GRRS, whatever their methods of detection, so that we can collectively describe ranavirus geography and host range.
Fig. 4.
Summary
Ranaviruses impact numerous ectothermic vertebrates of ecological and economic importance, as well as many of conservation concern (Chinchar and Waltzek 2014; Brunner et al. 2015). Tracking their geographic distribution and host range is fundamental to understanding their basic biology, epidemiology, and evolution and for efforts to mitigate their impacts. Long-term data sets also permit us to understand the effects that emergence has on populations. The GRRS thus represents an important resource for researchers and multiple stakeholders.
Our efforts to summarize the trends in the GRRS offer a perspective on the state of knowledge about ranavirus distributions. We find that there is growing number of occurrences of ranaviruses and a growing diversity of hosts. We expect this reflects a growing interest and study of ranaviruses, along with broader interest in host conservation (e.g., amphibian declines more broadly). We suspect it also mirrors the growing ease with which researchers can screen samples for ranaviruses and other pathogens. Indeed, the proportion of studies stemming from routine surveillance as opposed to investigating morbidity or mortality has grown alongside the huge increases in reports overall. We expect that the increasing use of environmental DNA (eDNA) for pathogen detection in a variety of settings (e.g., Hall et al. 2016; Vilaça et al. 2020) will further increase our understanding of at least the geographic distribution of ranavirus nucleic acids. We plan to modify the GRRS to allow for eDNA samples but encourage researchers to continue to screen hosts.
Our summary also makes clear that while records have increased dramatically, there still exist geographic and taxonomic biases. The vast majority of reports come from North America and thus North American species. Further sampling in South America, broader areas in Asia, and especially Africa will provide a much more complete picture of the geography of ranaviruses and their host ranges. Indeed, host ranges appears to be the most clearly biased aspect of our current sampling. One need only look to the severe under-representation of ranaviruses from fishes to appreciate the need for more surveillance in fishes. Additional sampling and genetic sequencing is also likely to add new viral species that may help elucidate phylogenies and evolutionary relationships of these important, emerging viruses.
We hope that as the GRRS becomes more widely known, more researchers will add their records and use it as a research tool. We have attempted to create a useful, open, free tool for ranavirus researchers, but in the end the GRRS will be as successful as the community makes it.
Acknowledgements
We are grateful for the hard work of many members of the Brunner lab, especially Ana Trejo, for a myriad of hours of data entry. We are pleased to acknowledge the support of the Wildlife Disease Association for a grant to support this data entry. Finally, the GRRS would not have been possible without the efforts of EcoHealth Alliance, especially Andrew Huff and Russell Horton, who created the initial version of the GRRS. This work was supported financially by the USDA Forest Service.
References
AmphibiaWeb. 2020. University of California, Berkeley, California [online]: Available from amphibiaweb.org/.
Blaustein A, Urbina J, Snyder P, Reynolds E, Dang T, Hoverman J, et al. 2018. Effects of emerging infectious diseases on amphibians: a review of experimental studies. Diversity, 10: 81.
Brunner J, Storfer A, Gray M, and Hoverman J. 2015. Ranavirus ecology and evolution: from epidemiology to extinction. In Ranaviruses: lethal pathogens of ecothermic vertebrates. Edited by MJ Gray and VG Chinchar. Springer International Publishing, Cham, Switzerland. pp. 71–104.
Chinchar VG, and Waltzek TB. 2014. Ranaviruses: not just for frogs. PLoS Pathogens, 10: e1003850.
Claytor SC, Subramaniam K, Landrau-Giovannetti N, Chinchar VG, Gray MJ, Miller DL, et al. 2017. Ranavirus phylogenomics: signatures of recombination and inversions among bullfrog ranaculture isolates. Virology, 511: 330–343.
Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, and Speare R. 1999. Emerging infectious diseases and amphibian population declines. Emerging Infectious Diseases, 5: 735–748.
Duffus A, and Olson D. 2011. The establishment of a global Ranavirus reporting system. FrogLog, 96: 37.
Duffus ALJ, Waltzek TB, Stöhr AC, Allender MC, Gotesman M, Whittington RJ, et al. 2015. Distribution and host range of ranaviruses. In Ranaviruses: lethal pathogens of ecothermic vertebrates. Edited by MJ Gray and VG Chinchar. Springer International Publishing, Cham, Switzerland. pp. 9–57.
Ebert D. 1998. Experimental evolution of parasites. Science, 282: 1432–1436.
Granoff A, Came PE, and Rafferty KA. 1965. The isolation and properties of viruses from Rana pipiens: their possible relationship to the renal adenocarcinoma of the leopard frog. Annals of the New York Academy of Sciences, 126: 237–255.
Hall E, Crespi E, Goldberg C, and Brunner J. 2016. Evaluating environmental DNA-based quantification of ranavirus infection in wood frog populations. Molecular Ecology Resources, 16: 423–433.
International Committee on Taxonomy of Viruses (ICTV). 2020. The ICTV report on virus classification and taxon nomenclature, Iridoviridae, Ranavirus [online]: Available from talk.ictvonline.org/ictv-reports/ictv_online_report/dsdna-viruses/w/iridoviridae/616/genus-ranavirus.
Rafferty KJ. 1965. The cultivation of inclusion-associated viruses from Lucke tumor frogs. Annals of the New York Academy of Sciences, 126: 3–21.
Schloegel LM, Daszak P, Cunningham AA, Speare R, and Hill B. 2010. Two amphibian diseases, chytridiomycosis and ranaviral disease, are now globally notifiable to the World Organization for Animal Health (OIE): an assessment. Diseases of Aquatic Organisms, 92: 101–108.
Smith K, Zambrana-Torrelio C, White A, Asmussen M, Machalaba C, Kennedy S, et al. 2017. Summarizing US wildlife trade with an eye toward assessing the risk of infectious disease introduction. EcoHealth, 14: 29–39.
Stagg H, Guðmundsdóttir S, Vendramin N, Ruane N, Sigurðardóttir H, Christiansen D, et al. 2020. Characterization of ranaviruses isolated from lumpfish Cyclopterus lumpus L. In the North Atlantic area: proposal for a new ranavirus species (European North Atlantic Ranavirus). Journal of General Virology, 101: 198–207.
Stöhr A, Fleck J, Mutschmann F, and Marschang R. 2013a. Ranavirus infection in a group of wild-caught Lake Urmia newts Neurergus crocatus imported from Iraq into Germany. Diseases of Aquatic Organisms, 103: 185–189.
Stöhr A, Hoffmann A, Papp T, Robert N, Pruvost N, Reyer H, et al. 2013b. Long-term study of an infection with ranaviruses in a group of edible frogs (Pelophylax kl. esculentus) and partial characterization of two viruses based on four genomic regions. The Veterinary Journal, 197: 238–244.
Tidona CA, Schnitzler P, Kehm R, and Darai G. 1998. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution. Virus Genes, 16: 59–66.
Vilaça ST, Grant SA, Beaty L, Brunetti CR, Congram M, Murray DL, et al. 2020. Detection of spatiotemporal variation in ranavirus distribution using eDNA. Environmental DNA, 2: 210–220.
Whittington R, and Chong R. 2007. Global trade in ornamental fish from an Australian perspective: the case for revised import risk analysis and management strategies. Preventive Veterinary Medicine, 81: 92–116.
World Organization for Animal Health (OIE). 2020. WAHIS portal: Animal Health Data [online]: Available from oie.int/en/animal-health-in-the-world/wahis-portal-animal-health-data/.
Information & Authors
Information
Published In

FACETS
Volume 6 • Number 1 • January 2021
Pages: 912 - 924
Editor: Ellen Ariel
History
Received: 28 February 2020
Accepted: 22 February 2021
Version of record online: 3 June 2021
Notes
This paper is part of a Collection titled “Ranavirus research: 10 years of global collaboration.”
Copyright
© 2021 Brunner et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
All conceived and designed the study.
JLB and ALJD performed the experiments/collected the data.
JLB analyzed and interpreted the data.
DLM contributed resources.
JLB, DHO, MJG, and ALJD drafted or revised the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Jesse L. Brunner, Deanna H. Olson, Matthew J. Gray, Debra L. Miller, and Amanda L.J. Duffus. 2021. Global patterns of ranavirus detections. FACETS.
6(): 912-924. https://doi.org/10.1139/facets-2020-0013
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Isolation, identification and the pathogenicity characterization of a Santee-Cooper ranavirus and its activation on immune responses in juvenile largemouth bass (Micropterus salmoides)
2. First evidence of ranavirus in native and invasive amphibians in Colombia
3. Is
Xenopus laevis
introduction linked with
Ranavirus
incursion, persistence and spread in Chile?
4. Non-Lethal Detection of Frog Virus 3-Like (RUK13) and Common Midwife Toad Virus-Like (PDE18) Ranaviruses in Two UK-Native Amphibian Species
5. Environmental and Anthropogenic Factors Shape the Skin Bacterial Communities of a Semi-Arid Amphibian Species
6. Comparison of detective ranavirus with major capsid protein gene from infected frogs (
Pelophylax nigromaculatus
and
Lithobates catesbeianus
) in South Korea
7. Prevalence of Ranavirus Infection in Three Anuran Species across South Korea
8. Ranavirus Amplification in Low-Diversity Amphibian Communities
9. Reptiles, Biodiversity of
10. Global Patterns of the Fungal Pathogen Batrachochytrium dendrobatidis Support Conservation Urgency
