Introduction
Anthropogenic stressors increasingly harm aquatic ecosystems, resulting in population declines of species over time. For the conservation of aquatic organisms, it thus becomes fundamental to understand how species and populations respond to a changing environment throughout their distribution. In their environments, organisms may experience abiotic stressors (e.g., temperature, dissolved oxygen, salinity, and pH), biotic stressors (e.g., intraspecific competition, invasive species, pathogens, and predators), and anthropogenic stressors (e.g., fisheries, habitat degradation, and pollutants) (
Schulte 2014). These environmental stressors may act on their own or, more likely, as parts of complex interactions (
Crain et al. 2008;
Jackson et al. 2016). Due to the presence of multiple stressors in many ecosystems, deciphering the responses of organisms to these environmental stressors can be challenging. Physiological responses of wild organisms are also mediated by previous exposure history as well as genomic variation that may be associated with adaptations to local environmental conditions (
Whitehead et al. 2011;
Crozier and Hutchings 2014). Therefore, when considering the conservation of a species, it becomes important to consider the physiological responses of that species across its distribution to determine how the local environment as well as genetic factors may influence these responses (
Homyack 2010;
Cooke et al. 2013).
The physiological response to stressors is typically characterized as an integrated response to cope with the stress and reestablish homeostasis. Part of this integrated organismal response to stressors is through changes in gene expression (e.g.,
Jeffries et al. 2021). The gene expression response to environmental stressors is complex and can be influenced by several factors, contributing to phenotypic variation within and among populations (
Whitehead and Crawford 2006;
López-Maury et al. 2008). The abundance of mRNA transcripts, the intermediate step between the transcription of the gene coding region of DNA and the translation into proteins, is a useful approximation of gene expression (
Buccitelli and Selbach 2020). In particular, whole-transcriptomic (e.g., mRNA sequencing (RNA-seq)) approaches provide a mechanism to examine transcriptome profiles of wild, non-model species, for which genomic resources may be limited (
Connon et al. 2018) to assess the cellular processes that are responding to a stressor. The transcriptomic responses of wild organisms to local environments can provide valuable insight into the cellular responses to a variety of stressors and are particularly useful when those environmental stressors are not well understood in a system (
Semeniuk et al. 2022). In line with this approach, transcriptomic analyses can provide information about what stressors may be present in an environment and acting on local species. For instance, liver transcriptional analyses revealed that silver carp (
Hypophthalmichthys molitrix (Valenciennes in Cuvier and Valenciennes, 1844)) residing at the leading edge of their range in the Illinois River (Illinois, USA) may be living in more polluted habitats compared to fish at the center of their distribution (
Jeffrey et al. 2019). Similarly, transcriptomic patterns in wild Pacific salmon suggest that responses to viral pathogens may be impacting both freshwater migration and return migration, as well as spawning success (
Miller et al. 2011;
Jeffries et al. 2014). Transcriptomics, thus, has the potential to provide invaluable information about possible environmental stressors and the physiological responses of organisms to these stressors.
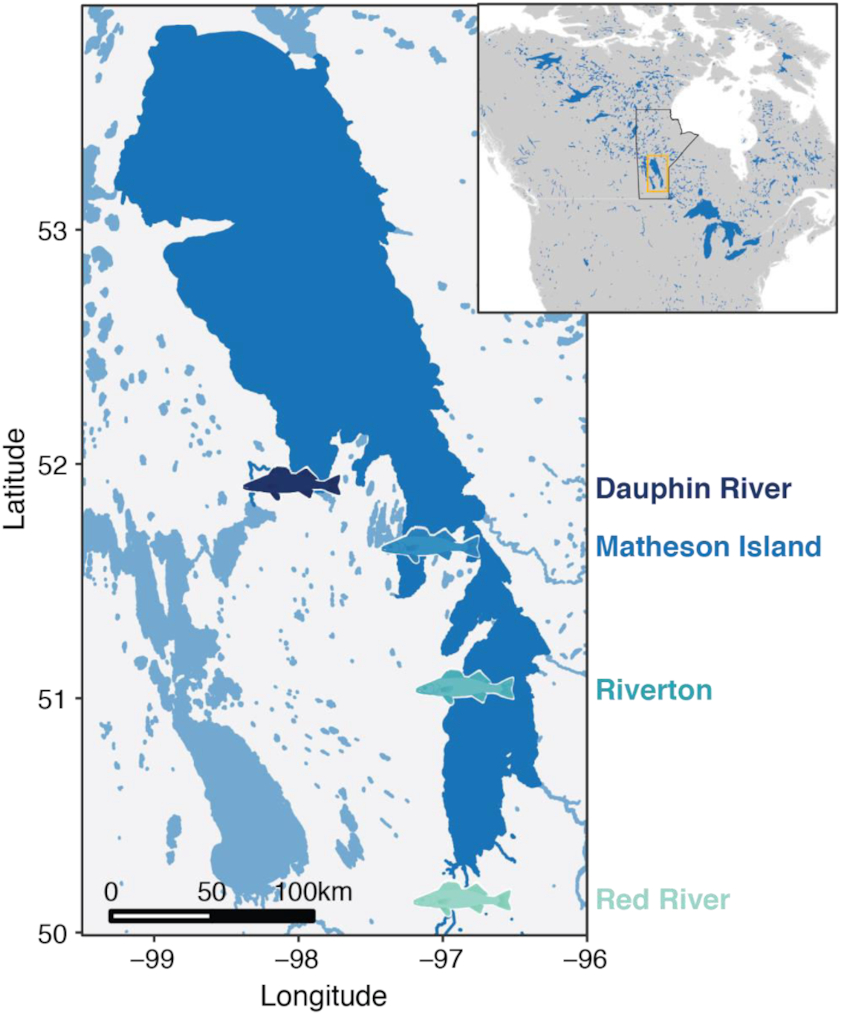
Freshwater fishes are some of the most at-risk organisms due to climate change as well as other human-related activities, such as fisheries. Lake Winnipeg (Manitoba, Canada) is the 10th largest lake globally based on surface area and is characterized by a large north basin, smaller south basin, and a narrow channel that connects the two basins (
Wassenaar and Rao 2012). Lake Winnipeg is home to Canada’s second largest freshwater fishery, of which walleye (
Sander vitreus (Mitchell, 1818)) make up the largest component (
Johnston et al. 2012). Sustainable management of walleye is thus critically important, not only to commercial fishing but also recreational angling, subsistence fishing, and the Lake Winnipeg ecosystem as a whole. Since 2002, however, the annual commercial yield from the walleye fishery has been above the maximum sustainable yield, and commercial harvests have declined between 2014 and 2018 (
Manitoba Government 2018;
Manitoba Sustainable Development 2018). The Lake Winnipeg ecosystem is simultaneously being impacted by additional stressors, such as eutrophication, invasive species, and collapse of a prey species (i.e., rainbow smelt;
Osmerus mordax (Mitchell, 1814)), that may be further exacerbating the fishery declines (
Schindler et al. 2012;
Jansen et al. 2017;
Enders 2019;
Enders et al. 2021;
Thorstensen et al. 2021). Reflecting this increase in anthropogenic pressure, walleye body condition has been declining across the lake (
Manitoba Government 2018;
Manitoba Sustainable Development 2018). Similarly, basin-specific effects are evident, with younger fish in the north basin showing smaller length-at-age, which may reflect nutritional limitation in more recent annual cohorts (
Thorstensen et al. 2021). A blood metabolite panel associated with amino acid metabolism and protein turnover in walleye also suggested that fish from the north basin (i.e., Dauphin River) may have elevated levels of protein degradation (
Thorstensen et al. 2021). Although low population divergence was detected for Lake Winnipeg walleye, sequence differences in mRNA transcripts associated with cell membrane protein and cytoskeletal function did vary latitudinally, suggesting that genomic differences may, in part, underlie phenotypic differences (
Thorstensen et al. 2020). Furthermore, contemporary gene flow from the connected Lake Manitoba (Manitoba, Canada) was concentrated in Lake Winnipeg’s north basin, possibly because of environmental differences between the basins of Lake Winnipeg (
Thorstensen et al. 2022). Together, the complexity of stressors potentially affecting walleye in the Lake Winnipeg system, as well as basin-specific genomic, physiological, and morphological metrics of walleye, suggest the need for conservation attention at a local scale.
The present study used the emerging challenges Lake Winnipeg walleye are facing to test whether whole transcriptome interrogation of individual fish from across regions can identify environmental stressors that might be contributing to population declines and morphological differences in walleye. To this end, walleye were sampled at locations representing the north basin (Dauphin River), the south basin (Red River), and the channel that connects the basins (Matheson Island) in 2017 and 2018. Gill samples were acquired non-lethally from walleye, allowing for the whole-transcriptome profiling of eight walleye from each location and year using RNA-seq. Due to the potential variability generated by sampling wild-caught fish, of particular interest were patterns of transcript regulation that were consistent across the two years examined. From these genes that differed in their mRNA abundance among locations consistently across years, a selection of 20 genes was examined using quantitative PCR (qPCR) across a larger sampling of fish, as well as from an additional site in the south basin (Riverton). Overall, by identifying consistent patterns of biological responses in walleye across the Lake Winnipeg system, information not only on walleye physiology but also potential environmental factors driving these responses can be determined. Furthermore, identification of key biological pathways or responses will facilitate targeted studies to draw more specific links between walleye genomic and phenotypic differences that may be driven by local environmental factors.
Discussion
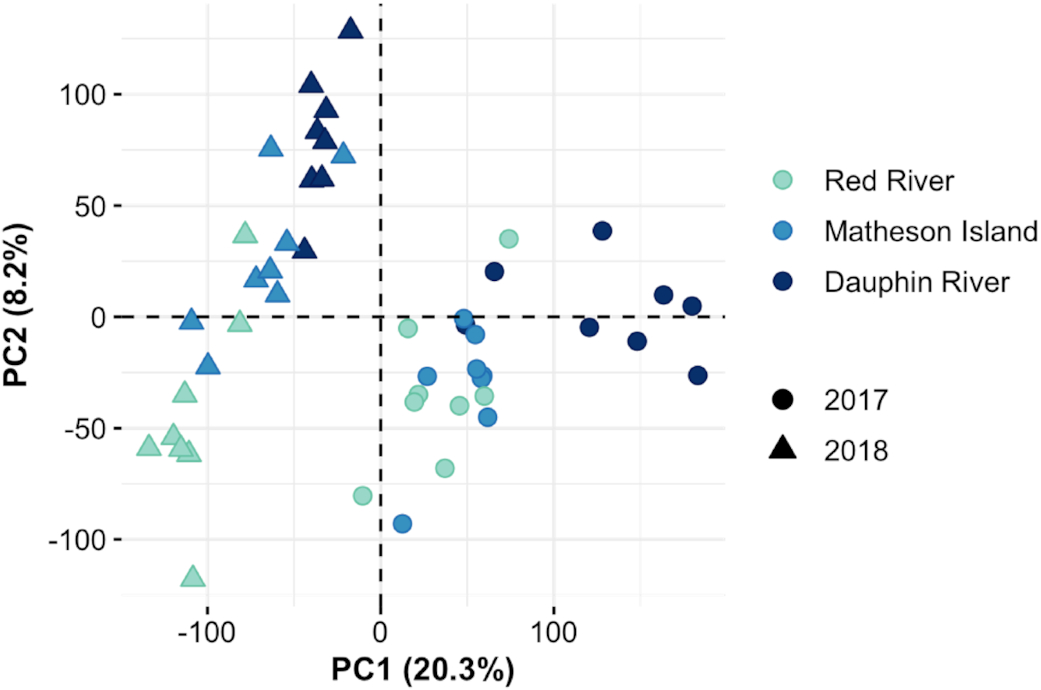
In the present study, RNA-seq was used to examine differences in the transcriptomic profiles of walleye that were non-lethally sampled at northern, central (channel), and southern locations within the Lake Winnipeg system. Since walleye were sampled from a wild setting with inherent environmental variability, of particular interest were differences in transcript levels that were consistent across the two years examined. Interestingly, several biological pathways/processes were consistently variable across the latitudinal gradient. In most cases, transcript levels of genes associated with a biological pathway were different between Dauphin River fish (northernmost site) and Red River fish (southernmost site), with the Matheson Island (channel) site being intermediate. Since no true “control” site was possible with the present study design, and to simplify discussion of these complex trends, we limited the discussion of the overall regulation of transcripts/pathways to those being highest or lowest in the northernmost (Dauphin River) compared to southernmost (Red River) locations.
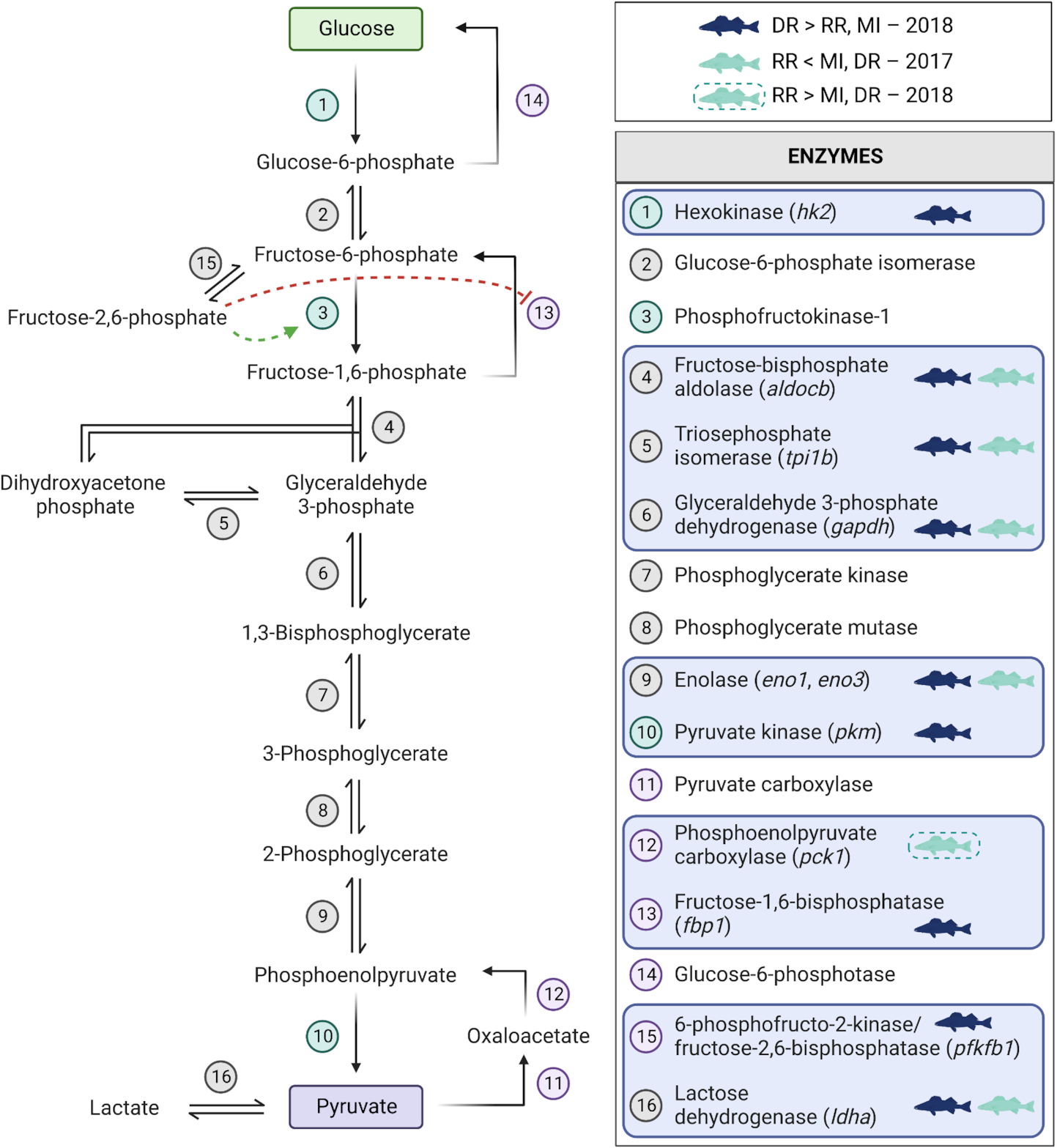
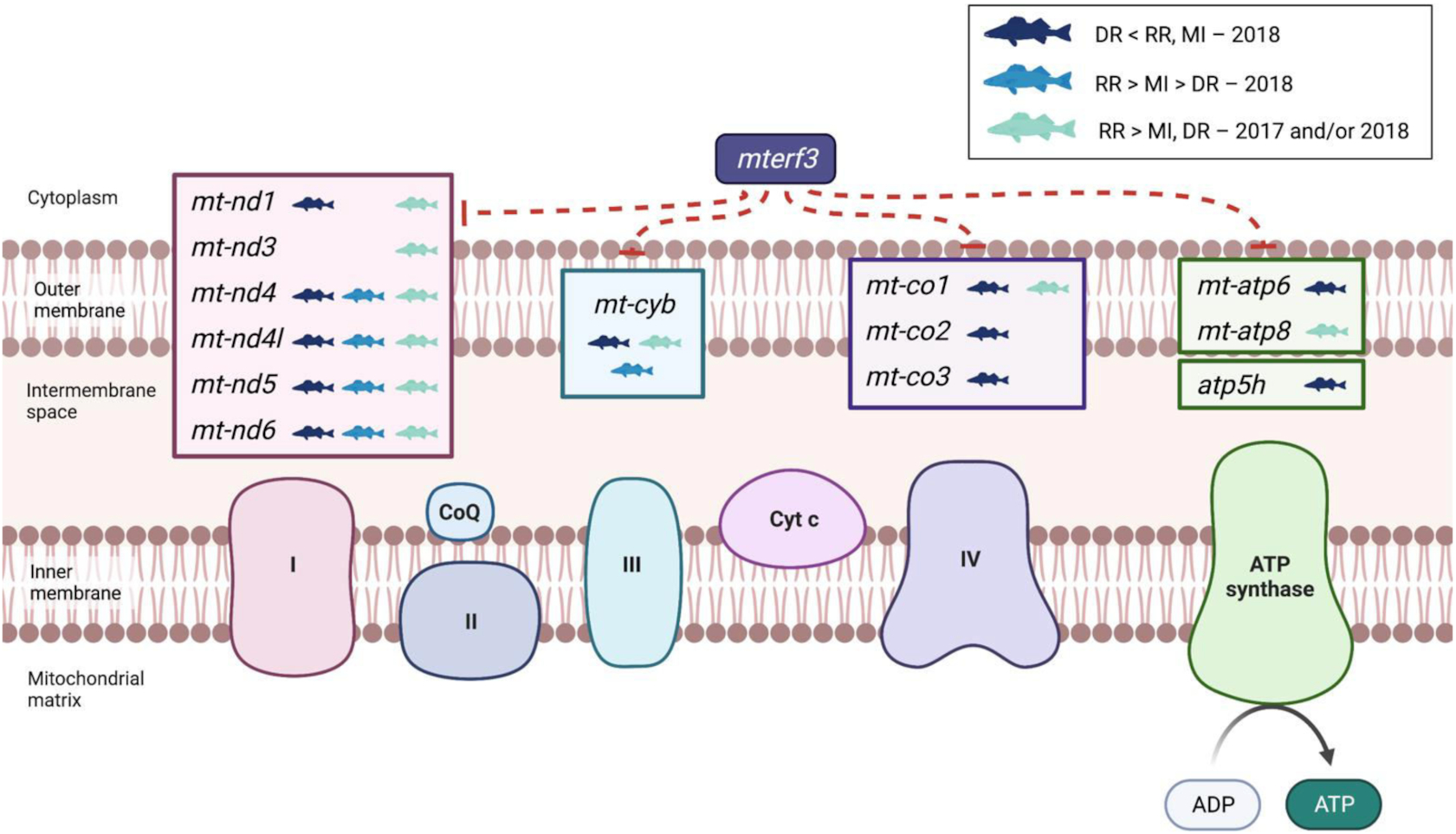
Walleye from the northernmost site exhibited elevated transcript abundances associated with a general response to low oxygen. For instance, transcript levels for several genes associated with glycolysis (e.g.,
fructose-bisphosphate aldolase C-B (
aldocb)glyceraldehyde-3-phosphate dehydrogenase (
gapdh),
alpha-enolase (
eno1)), as well as anaerobic metabolism (
ldha), were highest in Dauphin River fish consistently across years (
Fig. 3). A correspondingly lower level of transcripts for genes associated with the oxidative phosphorylation (e.g., genes related to complexes I, III, IV of the electron transport chain) was also observed in Dauphin River fish across both years (
Fig. 5). Interestingly, in 2018, the lower level of genes associated with the electron transport chain corresponded with elevated
mterf3 transcript levels, a negative regulator of mtDNA transcription in mammals (
Park et al. 2007). Collectively, these patterns suggest an elevated use of anaerobic pathways. Similar shifts in transcript regulation of metabolic processes in the gill have been detected in fishes exposed to both acute and chronic periods of hypoxia (
van der Meer et al. 2005;
Li et al. 2017). Further suggesting that walleye in the north basin may have encountered an increased frequency of low-oxygen conditions, hypoxia-related gene transcripts such as
hmox, hyou1, and
egln3 were highest in Dauphin River fish in 2017 or 2018, reflective of the potential activation of hypoxia-induced pathways (
Zhang et al. 2017). Although stratification of the Lake Winnipeg system is minor, the greater depth of the north basin (13.3 m in the north vs. 9.7 m in the south) results in a lower degree of mixing and may result in low-oxygen concentrations in the bottom waters of the north basin. Increasing levels of cultural eutrophication in Lake Winnipeg may also contribute to bottom water oxygen depletion (
Environment Canada and Manitoba Water Stewardship 2011). Oxygen-depleted waters (<2.9 mg L
−1) below the thermocline have been previously recorded across multiple years (2003, 2006, and 2007) in areas of the north basin during the summer months (
Environment Canada and Manitoba Water Stewardship 2011). Together, these data suggest regulation of genes involved in anaerobic metabolism and responses to hypoxia in north basin walleye, pointing to their potential exposure to low-oxygen environments.
Further indicators of general stress were observed in fish sampled from the north basin. Dauphin River fish exhibited the highest levels of mRNA transcripts associated with DNA repair and molecular chaperones (e.g.,
hsp90aa1 and
hsp90ab1 in 2018). Additionally, increased transcript levels of genes associated with proteasomal catabolism (e.g., proteasomal subunits
psm-d5,
d6) were observed consistently across years in fish from the Dauphin River. To support an increase in ammonium excretion across the gill as a result of protein breakdown, the mRNA abundance of several vacuolar-type H
+-ATPases was also consistently elevated in fish from the Dauphin River across years. Vacuolar-type H
+-ATPases are thought to support the production of an acidified gill water boundary layer to protonate excreted NH
3 (
Weihrauch et al. 2009). Walleye sampled in Dauphin River in 2017 showed elevated amino acid metabolites associated with post-transcriptional modifications of proteins, which may point to elevated turnover of previously synthesized proteins in these northernmost fish in comparison to the southernmost fish (
Thorstensen et al. 2021). Growth rates and body condition for Lake Winnipeg walleye have been declining in recent years, especially those from the north basin (
Thorstensen et al. 2021), results that echo the differences in mass and total length for fish sampled in the present study. These morphological differences are consistent with both blood metabolite and gill transcriptional results, suggesting increased endogenous protein breakdown and decreased nutritional status in fish from the north basin. Together, these data suggest that fish from the north basin may be experiencing increased exposure to environmental (e.g., oxygen) and nutritional stressors.
Transcript levels of genes associated with ion regulatory processes were also significantly altered among sampling sites and generally consistent across years; however, the direction of the response was gene-dependent. Transcripts of Na
+/K
+-ATPase 1 isoform
atp1a1 as well as Na
+/H
+ exchangers 1 and 3 (
slc9a1 and
slc9a3) were lowest in Dauphin River fish. Similarly, transcripts of several aquaporins and claudins were differentially regulated with most being lowest in Dauphin River fish. For ion regulatory gene transcripts that were highest for Dauphin River fish, the
atp1a3 isoform of Na
+/K
+-ATPase 1, calcium/calmodulin-dependent protein kinases (
camk-1, 1d, 2g), a tight-junction protein (
tjp3), and mineralocorticoid receptor (
nr3c2) were among those differentially regulated. Interestingly, genetic differences (e.g., single nucleotide polymorphisms (SNPs)) among Lake Winnipeg walleye were detected for transcripts related to ion channel activity (
Thorstensen et al. 2020). Multiple SNPs were identified in
atp1a3, Na
+/H
+ exchanger 6 (
slc9a6), and claudin-10 (
cldn10) transcripts that varied along a latitudinal gradient in Lake Winnipeg walleye (
Thorstensen et al. 2020). Salinity and specific conductance differ across the Lake Winnipeg system, with higher levels being detected in the north basin (Na
+ and Cl
− 17–19 mg L
−1; 313 µS cm
−1) compared to the south basin and channel (Na
+ and Cl
− 9–11 mg L
−1; 287 µS cm
−1) (
Environment Canada and Manitoba Water Stewardship 2011). Spatial differences in specific conductance and salinity are likely driven by the Dauphin River that transports ion-rich waters (Na
+ and Cl
− 184–305 mg L
−1; 1200–1600 µs cm
−1) from the more saline Lake Manitoba into Lake Winnipeg (
Environment Canada and Manitoba Water Stewardship 2011). Lower salinity and specific conductance inflows from the Winnipeg River (Na
+ and Cl
− 1–2 mg L
−1; 100 µS cm
−1) also help to dilute waters in the south basin and channel (
Environment Canada and Manitoba Water Stewardship 2011). Spatial variation in other physicochemical properties, such as sulfate, phosphorus, nitrogen, suspended solids, as well as temperature, precipitation, and river discharge, may also contribute to the potential ion regulatory differences in Lake Winnipeg walleye (
Environment Canada and Manitoba Water Stewardship 2011). Together, these results suggest a level of reorganization of ion regulatory machinery that is sensitive to local conditions; however, the underlying mechanisms remain unclear.
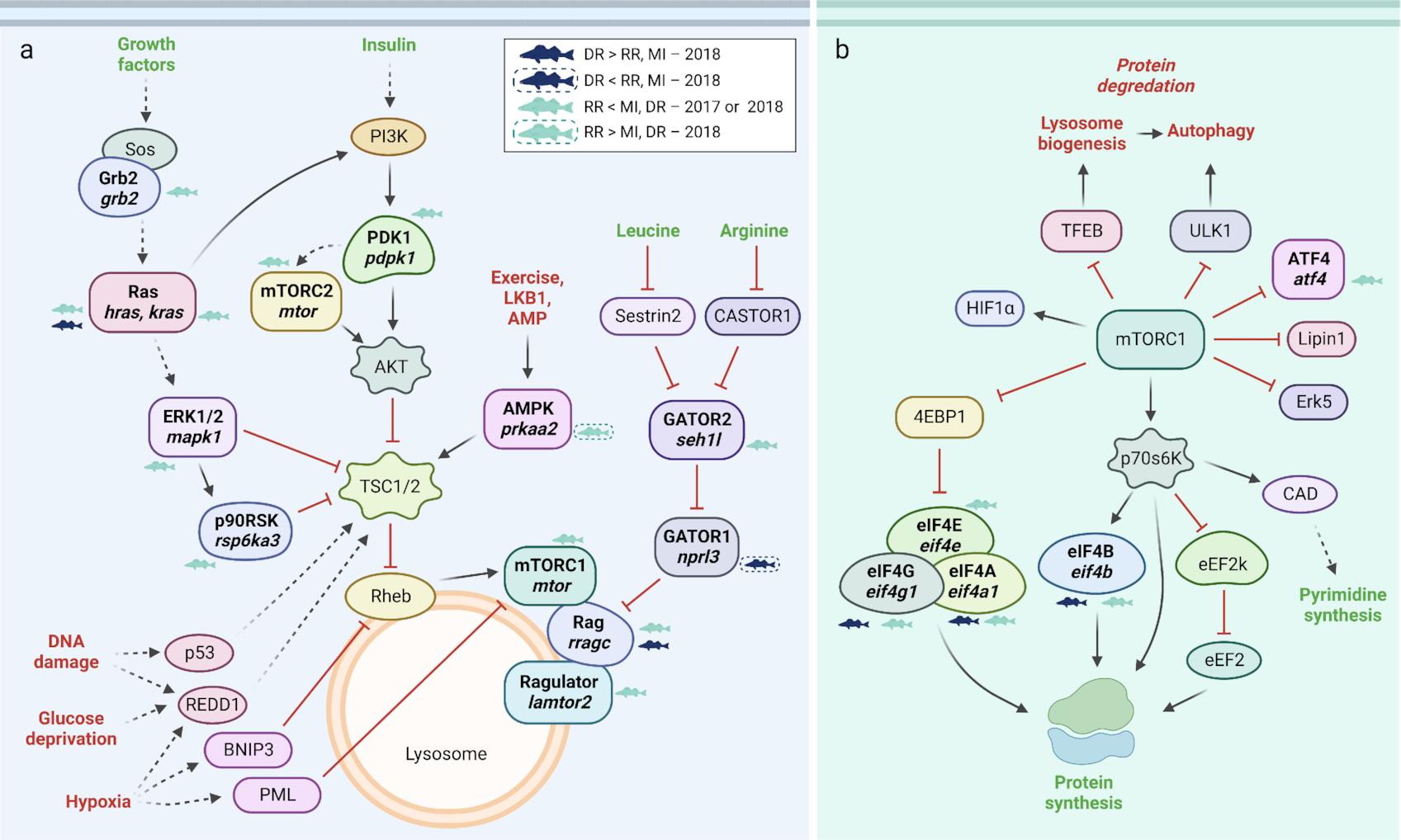
In line with a potential gill ion regulatory response, aspects contributing to possible gill remodeling were also evident at a local scale. Several genes associated with the mTOR pathway were identified as differentially regulated across sites with some of these genes being consistently regulated across years (
Fig. 4). Overall, activation of the mTOR pathway seemed evident for fish from the Dauphin River, in addition to downstream processes, such as increased protein synthesis and processing, as well as decreased autophagy. Similarly, key regulators and targets of AKT and the PI3K/AKT/mTOR pathway were highest for Dauphin River fish. For instance,
mtor and
pdpk1 that are involved in the activation of AKT were highest in fish from the north basin, as well as genes involved in the targeted effects of AKT such as fatty acid synthesis (
acly, acaca) and cell survival and proliferation (
foxo1a). Additionally, enrichment of biological processes such as actin cytoskeleton reorganization suggests potential alterations to the cellular and structural makeup of the gill architecture/tissue of fish from the north basin (
Ciano‐Oliveira et al. 2006;
Fiol et al. 2006). Similarly, transcripts with multiple SNPs related to the cytoskeleton varied along the latitudinal gradient in Lake Winnipeg walleye, suggesting a possible underlying genetic difference in these structural transcripts driven by local conditions (
Thorstensen et al. 2020). Together with the responses indicative of exposure to environmental stressors in the Dauphin River fish, regulation of transcripts involved in remodeling of the gill may reflect differences in local environmental conditions (e.g., potential differences in oxygen or salinity).
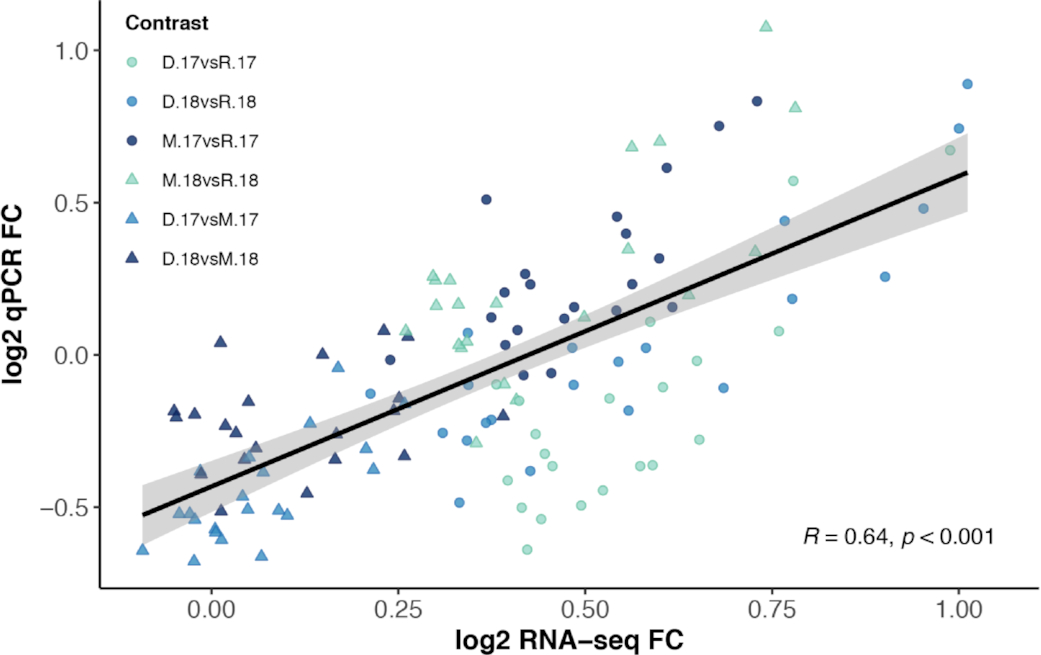
Transcriptomic responses of walleye in the system were verified in two ways in the present study. First, 20 target genes were identified using a PCA based on their differential regulation across sampling sites and examined using qPCR. Log2 fold change transcript levels were significantly correlated between qPCR and RNA-seq platforms. Similarly, examining the relative mRNA levels using a larger sample size suggested that the fish used for the RNA-seq study were representative of a larger group of fish for all but 1 of the 20 target genes. Secondly, relative mRNA levels were examined using qPCR across four sampling sites, which included the addition of the Riverton location in the south basin that was not included in the RNA-seq study. Even with significant year effects for several of the genes of interest, location-specific effects were evident for 10 of the 20 genes that were consistent with the RNA-seq results. For instance, transcript levels of gpx1 and rpa2 involved in the stress response and DNA repair, respectively, as well as cotl1, actr2, fam49b, and pfn2 involved in cytoskeleton organization, were all consistently lowest in Red River fish across years (Fig. S9). Together, these results suggest consistency among the RNA-seq and qPCR platforms as well as identify potential markers of stress responses and cytoskeleton reorganization in the gill that could be further explored for differences at a local scale among walleye in Lake Winnipeg or other systems.
With concerns for the status of the walleye fishery in the Lake Winnipeg system, a greater understanding of the state of this ecosystem is of vital importance. The generation of tools to improve our underlying knowledge of the physiology of walleye can improve our understanding of their condition and provide tools to monitor their status in the wild. Traditional measures of fisheries assessment, such as growth and reproductive outputs, are useful in providing population-scale indices, but these metrics are often based on terminal sampling and only evident at the point where vast changes to the population have already begun to take place (
Attrill and Depledge 1997;
Jeffrey et al. 2015). Using molecular and physiological tools through non-lethal sampling, as was done in the present study, has the potential to provide information about the health and status of a population on a shorter time scale, to complement population-level fisheries assessments (
Jeffrey et al. 2015,
2020;
Jeffries et al. 2021;
Semeniuk et al. 2022). In the present study, gill transcriptomic patterns suggested that location-specific environmental factors (potentially dissolved oxygen and salinity/specific conductance levels) may be present and driving changes in the gill physiology of fish in the north basin compared to those of the south basin. Using qPCR and a targeted list of genes, we verified differential regulation of many of these pathways by location, as well as identified potential targets that could be used for assessing walleye physiology in the future.
Gill tissue provides an excellent target for studies that wish to non-lethally sample fish. From a physiological perspective, gills are a multi-functional tissue that are at the interface of the internal and external environments and are sensitive to environmental and biological stressors (
Jeffries et al. 2021). Gill biopsies can also be retrieved with relative ease and with limited long-term effects on the fish, having been shown to have no subsequent impact on growth or survival (e.g.,
Martinelli-Liedtke et al. 1999;
Zhao et al. 2014). Gill biopsies are being increasingly used to monitor fish health, with the gill transcriptome having been shown to be sensitive to a variety of stressors (e.g., environmental conditions on migration and spawning failure (
Evans et al. 2011;
Miller et al. 2011), pathogens (
Jeffries et al. 2014), and elevated temperature and hypoxia (
Jeffrey et al. 2020)). While results from the present study and those of previous studies reflect the value of using non-lethal gill samples to examine fish health, tissue-specific transcriptional patterns may limit our ability to capture “the whole picture” with a single tissue (
Jeffries et al. 2021). For instance, using gill tissue may bias the potential environmental stressors identified in this study as those most likely to be exhibited by the gill transcriptome (e.g., temperature, dissolved oxygen, and ionic disturbances), whereas liver samples may have revealed additional information regarding toxicological stressors or the head kidney would provide valuable information on immune responses. Furthermore, sex-specific differences in walleye transcriptomic patterns were not examined in the present study due to limitations in accurately determining sex visually in this monomorphic species. However, when individuals of unknown sex were removed from the candidate gene mRNA dataset, there was no significant effect of sex when included as a covariate on the 20 genes evaluated. This limited analysis of gill tissue for sex-specific effects in the present study does not suggest an absence of sex effects in the fish sampled but may point to another tissue (e.g., liver and gonads) as being more suitable to examine such differences in wild-caught fish. Therefore, it is necessary to consider that walleye in the Lake Winnipeg system may exhibit additional responses to stressors not identified in the present study using gill tissue, but that data from the present study can be used to “spring-board” future work.
As conservation challenges threaten fish populations worldwide (
Arthington et al. 2016;
Dudgeon 2019), physiological responses can contribute to addressing those challenges by informing management and policy decisions (
Somero et al. 2016). Transcriptomics can contribute to physiological information by measuring responses to stressors that may or may not be known in advance of sampling (
Jeffries et al. 2021). Therefore, transcriptomic approaches may be useful for assessing physiological responses in wild organisms and contributing to management decisions (
Connon et al. 2018). This study outlines an approach to using transcriptomics to non-lethally describe stress responses in a wild fish. We used mRNA sequencing to observe overall patterns in gill gene expression changes and qPCR to test hypotheses generated with mRNA sequencing and elucidate the population-scale patterns of specific genes by broadening the sample set used. Similar approaches, as well as the targets identified in this study, could be applied in other systems where undefined local conditions may be suspected as contributing to fish health and status. Overall, the results of the present study highlight the potential for incorporating high-throughput molecular techniques in the conservation of non-model species.